Primarily screening and analyzing ESTs differentially expressed in rats’ primary liver cancer
Introduction
The development of primary liver cancer is a complicated procedure involving multiple factors and polygene mutations. Its incidence and mortality rank second among all cancers in China. It is often symptomless, usually occult in onset, and has nonspecific symptoms even in part of advanced liver cancer, so it is difficult to find out whether a person has liver cancer or not in very early stage (1). About 10% of primary liver cancer comes from the malignancy of biliary epithelium, indicating that the biliary epithelium has a greater potential to develop into cancer than hepatocytes. Moreover, intrahepatic cholangiocarcinoma (ICC) has been described as being difficult to distinguish from hepatocellular carcinoma (HCC) and exhibits a pseudoglandular or poorly differentiated morphology, so the rate of early detection and diagnosis is greatly reduced (2). Although, it has been reported that ICC is associated with the inactivation of certain tumor suppressor genes such as p53, adenomatous polyposis coli (APC) and smad-4 as well as mutations in oncogenes such as K-ras, c-myc and c-erb-2 (3), the mechanisms related to the overall process remain largely unknown. In addition, the present studies mainly focused on the molecular mechanism of hepatic cancer, and most of the experimental materials were obtained from clinical patients with moderate and advanced liver cancer. As a result, that greatly limited the systematic studies of molecular mechanisms in early stage of canceration, and the comprehensive analyses were lacked at gene level during the carcinogenesis.
Express sequence tags (ESTs) have also been found to be useful for screening new unknown genes, and the differential and quantitative analysis of expression patterns and for the evaluation of gene expression profiles in cancers. The analysis using EST sequences has an advantage of selecting new candidate genes (2). In a previous study, using ESTs, we identified some novel genes that offered valuable information related to the development of NIH mice’s fore stomach carcinoma (4).
In this study, the SD rat liver cancer model, the pathological changes of which were much similar to human primary liver cancer, was built by diethylnitrosamine (DENA) gavage, then mRNA differential display reverse transcription PCR (DDRT-PCR) was used to primarily screen differentially expressed ESTs. The aim of this study was to study the related genes in carcinogenesis and the development of liver cancer, discuss the molecular mechanisms, and provide the important theory basis and references for tumor clinical early diagnosis and therapies.
Materials and methods
Experimental animals
Ninety male SD rats (SPF III, 112±25 g, 4 weeks old) were purchased from the Experimental Animal Center of Henan Province (Zhengzhou), and randomly divided into two groups: the induced group (70 rats) and the control group (20 rats). The animal experiments were approved by the Institutional Animal Care and Use Committee of the College of Life Science, Henan Normal University.
Inducing drugs and primers
DENA was purchased from Sigma (St. Louis, USA). RNase-free DNase I and TrizolTM reagent were respectively bought from Promega (Madison, USA) and Tiangen (Beijing, China). RevertAidTM First Strand cDNA Synthesis Kit was purchased from Fermentas (Berlington, Canada). Taq DNA polymerase, dNTPs and pMD18-T Vector were all purchased from TaKaRa (Dalian, China). DNA gel purification kit was bought from SunBiotech (Beijing, China).
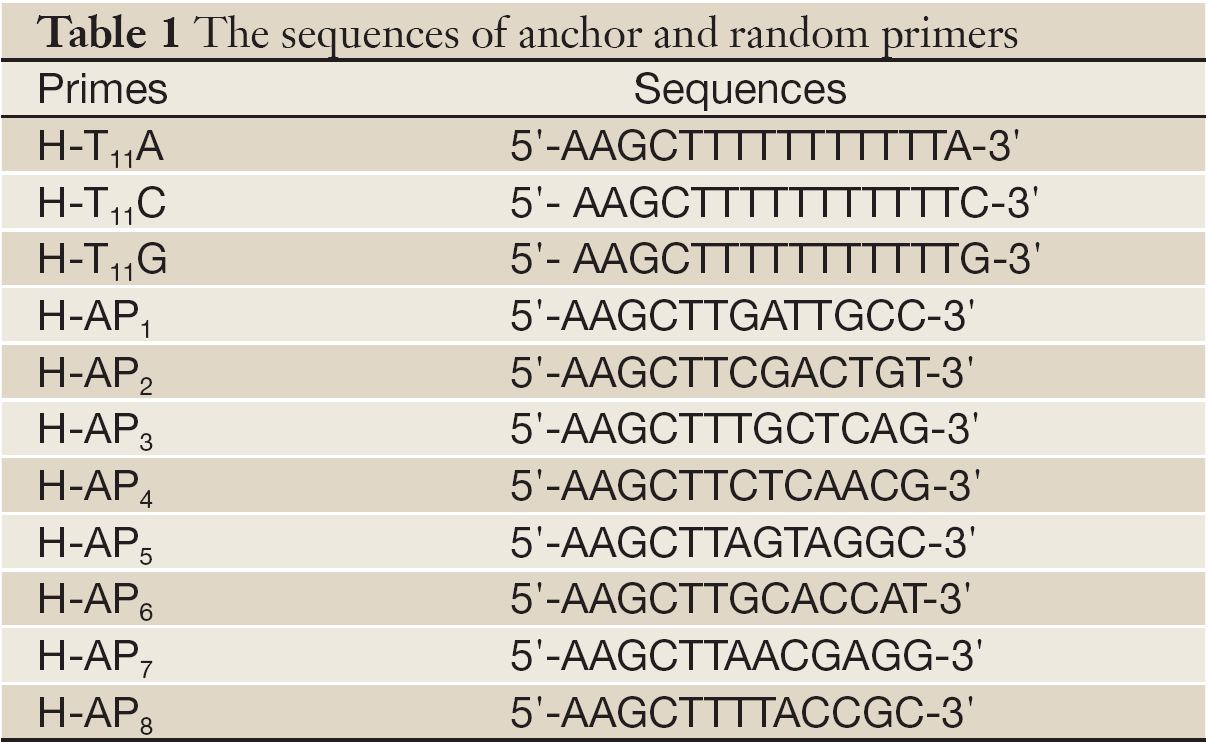
The third generation primer sequences designed by Liang et al. were used as degenerate primers. There were 3 down anchor primers and 8 up random primers. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers: 5'-GTGATGCTGGTGCTGAGTATGTC-3'and 5'-CAGTCTTCTGAGTGGCAGTGATG-3'. Primers were all synthesized by Shanghai Sangon (Shanghai, China).
Establishment of animal models
In the induced group, SD rats’ hepatocarcinoma was induced by gavage method. 1% DENA aqueous solution was administered at the dose of 70 mg/kg bw once a week for 15 weeks, then on normal diet. The control group was treated by gavage as the same dose of 0.9% physiological saline. Based on the relevent rules and guidelines about the experimental animals in China, cervical dislocation as the most humane way was used in this study. Rats were sacrificed by six times at d 14, d 28, d 56, d 77, d 105 and d 112 respectively. Then rats’ livers were made into paraffin sections, stained by hematoxylin-eosin (HE) and observed in different pathological periods by two senior pathologists of medical college.
RNA isolation and purification
Total RNA was extracted with the recommendations of the Trizol Regent manufacturer. After measuring OD260/OD280, 1.5% agarose gel electrophoresis was used to test whether RNA was degraded, then total RNA was treated by RNase-free DNase I to get rid of genome DNA.
Reverse transcription and cDNA identification
First-strand DNA was prepared from pure total RNA using 3 different down anchor primers respectively and RevertAidTM first strand cDNA synthesis kit. GAPDH, house keeping gene, was amplified by PCR with cDNA as templates while the quality of cDNA was tested.
PCR amplification
The PCR reactions were performed using all combinations of one random primer and one anchor primer (Table 1). cDNA was diluted to 1 mg/µL, and PCR was performed in 20 mL reaction system (1× reaction buffer, 1.5 mmol/L MgCl2, 0.1 mmol/L dNTPs, 1 mmol/L anchor primer, 0.2 mmol/L random primer, 1 mL cDNA and 1U Taq polymerase) for 1 cycle (94 °C, 5 min), 5 cycles (94 °C 30 s, 40 °C 2 min, 72 °C 30 s), 35 cycles (94 °C 30 s, 55 °C 2 min, 72 °C 30 s) and 1 cycle (72 °C, 7 min).
Full table
Denaturized polyacrylamide gel electrophoresis (PAGE) and secondary PCR
The PCR products (15 µL) were denatured at 95 °C for 5 min and 6% denaturized PAGE was carried out. Then, differential DNA bands were displayed by silver staining method. After comparison and analysis, differential bands were cut from the gel and placed for more than 1 h at 37 °C. After centrifugation, supernatants were used to amplify with the same primer set and conditions, but 0.2 mmol/L dNTPs substituted for 0.1 mmol/L dNTPs. The secondary PCR products were separated with 1.5% agarose gel electrophoresis, purified and retrieved.
Cloning and sequencing
The secondary PCR products were cloned into pMD18-T vector and transformed into E. coli DH-5α. The fragments were sequenced by Beijing SunBiotech.
Bioinformatics analysis
BLASTN 2.2.18 program was used for homologous comparison in GENBANK, and the information of the related homologous ESTs could be obtained. Eventually, the sequences and related proteins were analyzed.
Results
Pathological classification
According to the histological and cytological changes in hepatocarcinogenesis (5,6), the rat livers from animal models could be divided into the following types: normal (N), light inflammation (LI), heavy inflammation (HI), nodus hyperplasia (NH), untypical nodus hyperplasia (UNH) and ICC.
RNA isolation
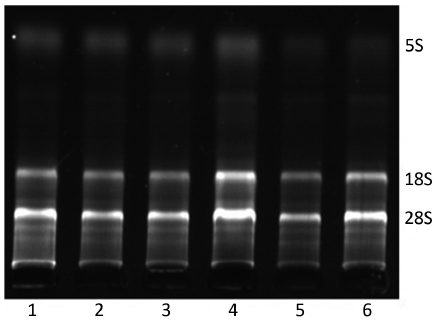
The bands of 28S and 18S of total RNA were all clear while the ratio of 28S/18S was 2:1, and there was no protein contamination (Figure 1). Furthermore, OD260/OD280 was 1.8-2.0. The results showed the extracted total RNA was intact and had high purity.
cDNA identification
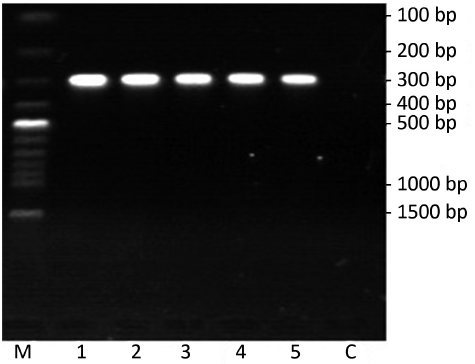
In electrophoresis picture, one bright and clear band about 300 bp showed that successful reverse transcription and cDNA had high quality (Figure 2). There was no band in lane C with RNA as the template. It suggested RNA was not contaminated by genome DNA.
Differential expressed cDNA isolation and reamplification
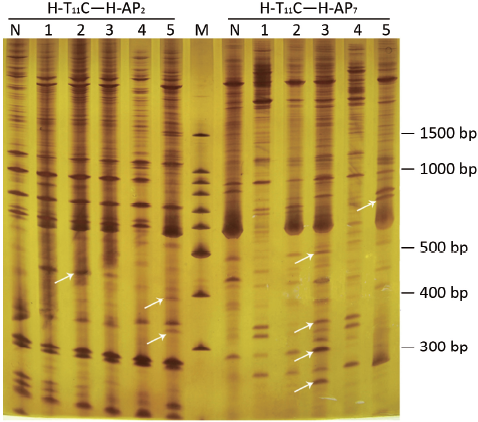
In Figure 3, the comparison showed that the abundance of certain DNA bands between normal and pathological stages was displayed in a pattern of up-regulation, down-regulation, generation or loss. Among them, 101 differential expressed bands were targeted. After retrieving and re-amplification, 55 differential expressed bands were obtained eventually (Figure 4).
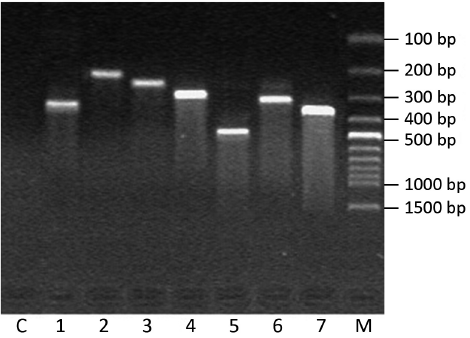
DNA sequencing and bioinformatic analysis
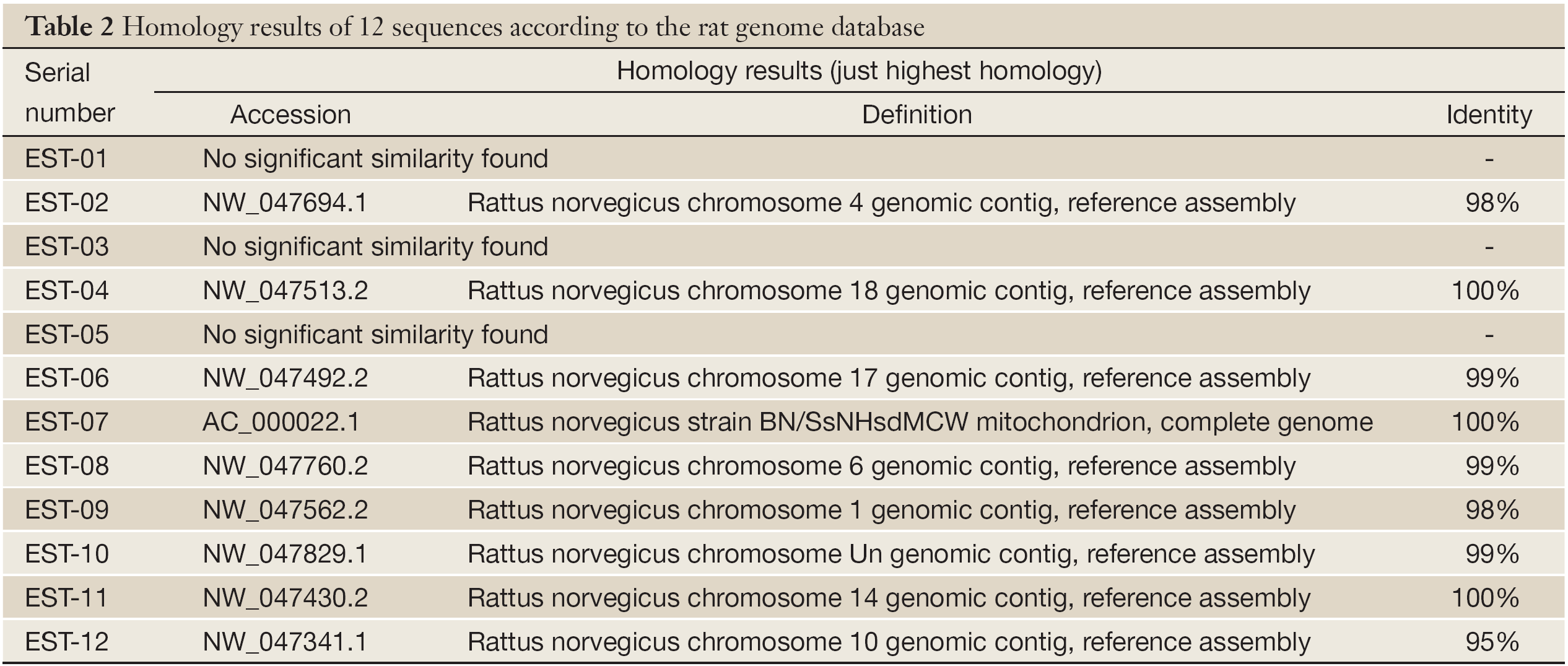
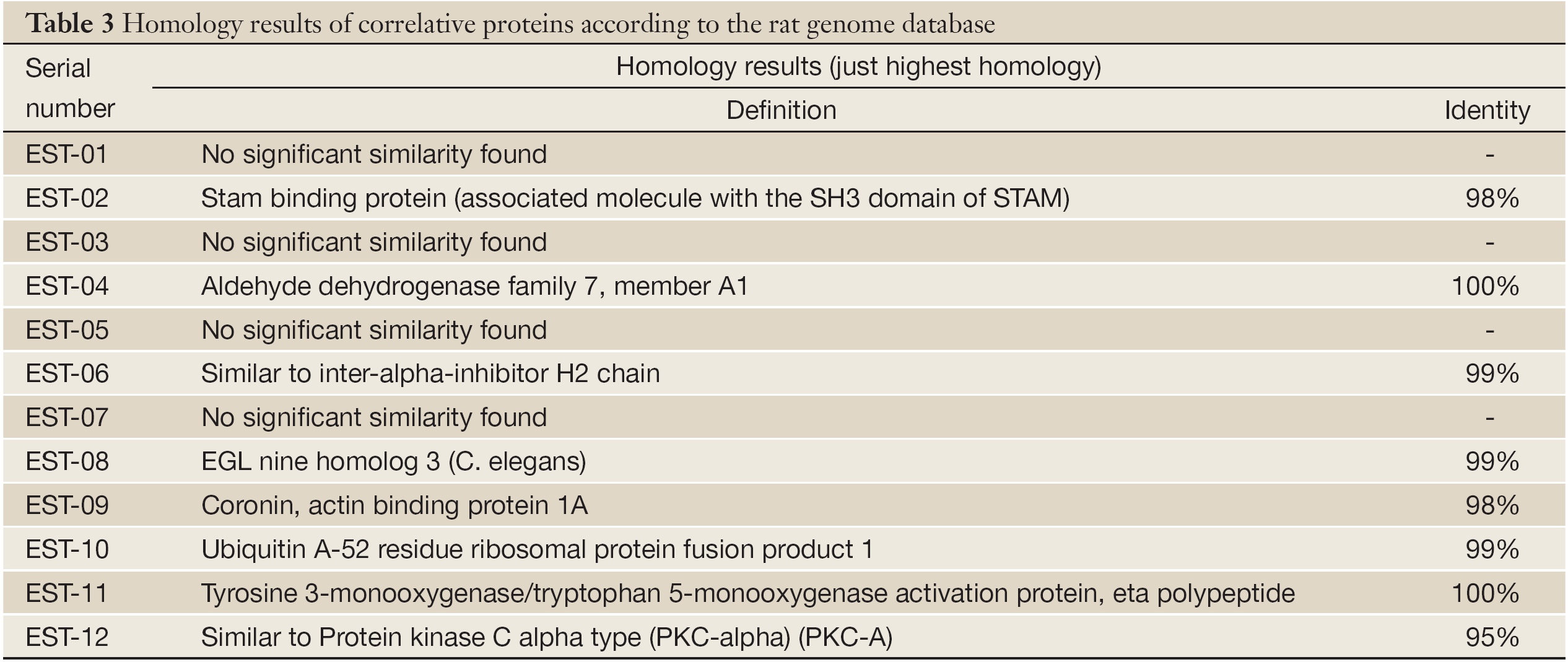
Twelve genes were transformed into E. coli DH-5α and sequenced. After homological analysis, 3 fragments were new unknown genes and 9 were known genes. Among them, EST-7 was identical to rat mitochondria gene BN/SsNHsdMCW, and the encoded protein was unknown (Tables 2,3).
Full table
Full table
Discussion
Liver cancer consists of three types: HCC, ICC and combined hepatocellular and cholangiocarcinoma. The incidence of HCC is the highest and 15% of liver cancer cases all over the world are ICC (7). As one subtype of liver cancer, researches and reports about ICC are still rare either in domestic or foreign on its clinical diagnosis and molecular mechanism (8-10).
Among the twelve differential expressed fragments obtained, EST-7 is highly homologous (100%) with rat mitochondria gene BN/SsNHsdMCW and considered as the part of BN/SsNHsdMCW sequence. In 2006, Li (11) was studying rats with cardiomyopathy and found that mitochondria gene BN/SsNHsdMCW was up-regulated after bone marrow mesenchymal stem cell transplantation. BN/SsNHsdMCW is considered to be relevant to mitochondria synthesis. When it and other related genes are up-regulated, numbers of mitochondria increase and cardiac function would be improved. There are multiple variants of mitochondria gene in rat genetic models of hypertension, the lack of any sequence variants provides conclusive evidence that divergence in blood pressure is exclusively programmed through their nuclear genomes (12). Moreover, variants of mitochondria gene are closely related with type I and II diabetes (13-15). Mitochondria have play a crucial role in carcinogenesis and development of many cancers (16-20), but most of the present researches are focusing on variants or the instability of mitochondria genes, and few direct evidence between BN/SsNHsdMCW and carcinogenesis is available.
In this study, EST-7 is expressed highly in normal livers, down-regulated in LI, and up-regulated in HI and NH. Although its expression declines again in UNH, it is highly expressed in ICC. It shows that liver cell malignant transformation is closely related to complex changes of mitochondria synthesis which may directly provide the energy for hepatocarcinogenesis. In early stage, the toxicity of DENA could activate the rat’s stress reaction, mitochondria synthesis decreases as well as energy metabolism. Finally, that might cause low expression of EST-7 in LI. In HI, more cells are malignant and more energy is required for rapid growth, so synthesis and numbers of mitochondria must be increased to promote the energy metabolism by up-regulating EST-7. With serious lesion, cachexia induced by liver evil proliferation seriously interferes in the normal metabolism of the body, so the mitochondria synthesis and energy metabolism are all decreased in UNH. In advanced stage, tumor growth, invasion and metastasis require more mitochondria to generate more energy. Tumor cells could up-regulate the synthesis genes to supply adequate energy for immortal growth. The protein encoded by BN/SsNHsdMCW and its mechanism should be further studied in detail.
In addition, EST-7 is also completely homologous with Ac2047 which is down-regulated in rat liver regeneration. It suggests that EST-7 may play certain role in the stress reaction, division and proliferation of liver cells, and mitochondria function may be also promoted by this gene to generate more energy for cell proliferation.
After blasting rat genome database, EST-12 is homologous (95%) with PCK-alpha on chromosome 10. Khan (21) found down-regulated PCK-alpha may be very important in carcinogenesis involved with Ras mutation. When Bianchi (22) researched on distribution of T-cell signaling molecules in human myeloma, he found the expression of PCK-alpha declined greatly. In this study, down-regulation of EST-12 in every stage further shows PCK-alpha is closely related to carcinogenesis. It is speculated that PCK-alpha may change the activities of regulation proteins by regulating some tumor suppressor genes. Down-regulation of PCK-alpha may cause abnormal phosphorylation of regulation proteins, and the related suppressor genes are restrained and not activated to transform. At last, the negative control of cell proliferation is lost and that can promote carcinogenesis with simultaneous ras mutation (23-25). But the mechanisms and genes regulated by PCK-alpha have not been elucidated.
The related proteins of other seven ESTs are respectively associated molecule with the SH3 domain of STAM (AMSH), aldehyde dehydrogenase family 7, inter-alpha-inhibitor, EGLN3, coronin and actin binding protein 1A, ubiquitin A-52 residue ribosomal protein fusion product 1 and tyrosine 3/tryptophan 5-monooxygenase activation protein (Table 3). They are closely related to the carcinogenesis, invasion and metastasis. For example, AMSH and tyrosine 3/tryptophan 5-monooxygenase activation protein are kinases/regulatory factors which take part in cell signal transduction pathways by “on-and-off” switching other factors of the pathway, then promotive and inhibitive signals of cell growth are transmitted into cell nucleus to activate or restrain transcription of genes bound up with cell growth and proliferation, thus cell dedifferentiation and malignant transformation are interfered by them (26,27). Inter-alpha-inhibitor is an important factor in cell apoptosis. Its abnormal expression would make tumor cells escape from cell death program and immune surveillance for immortal division and proliferation. Aldehyde dehydrogenase family 7 is a member of cell metabolism enzyme families. During carcinogenesis, its normal characteristics will be changes, and that inevitably results in metabolism chaos and abnormal expressions of related proteins which could feed back to physiological functions of other cells (28-31). Moreover, coronin and actin binding protein 1A is related to organize cytoskeleton. It can keep or change the stabilities of cytoskeleton by changing actin conformations, induce cell morphological and biochemical changes and participate in carcinogenesis, invasion and metastasis (32-34).
Since EST-1, EST-3 and EST-5 have no homology with any known genes, they are considered as new genes. Their whole sequences, conformations of related proteins and functions in tumors will need more researches in the future.
Acknowledgements
This work was supported by the Key Program for Science and Technology Development of Henan Province [122102310174], and the Zoology Key Subject of Henan Province.
Disclosure: The authors declare no conflicts of interest.
References
- Chuang SC, La Vecchia C, Boffetta P. Liver cancer: descriptive epidemiology and risk factors other than HBV and HCV infection. Cancer Lett 2009;286:9-14.
- Wang AG, Yoon SY, Oh JH, et al. Identification of intrahepatic cholangiocarcinoma related genes by comparison with normal liver tissues using expressed sequence tags. Biochem Biophys Res Commun 2006;345:1022-32.
- Shirabe K, Shimada M, Harimoto N, et al. Intrahepatic cholangiocarcinoma: its mode of spreading and therapeutic modalities. Surgery 2002;131:S159-64.
- Zhang HX, Liu DD, Li L, et al. Screening and identification differentially displayed genes in fore stomach carcinoma of mice. Chin J Cancer Res 2009;21:37-43.
- Histologic typing of liver tumors of the rat. Institute of Laboratory Animal Resources, National Research Council, National Academy of Sciences, Washington, D.C. J Natl Cancer Inst 1980;64:177-206.
- van Malenstein H, van Pelt J, Verslype C. Molecular classification of hepatocellular carcinoma anno 2011. Eur J Cancer 2011;47:1789-97.
- Olnes MJ, Erlich R. A review and update on cholangiocarcinoma. Oncology 2004;66:167-79.
- Kanamoto M, Yoshizumi T, Ikegami T, et al. Cholangiolocellular carcinoma containing hepatocellular carcinoma and cholangiocellular carcinoma, extremely rare tumor of the liver:a case report. J Med Invest 2008;55:161-5.
- Xu X, Kobayashi S, Qiao W, et al. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J Clin Invest 2006;116:1843-52.
- Sempoux C, Jibara G, Ward SC, et al. Intrahepatic cholangiocarcinoma: new insights in pathology. Semin Liver Dis 2011;31:49-60.
- Li Y, Chen Y, Tian J, et al. Screening of differentially expressed genes in rats with cardiomyopathy after bone marrow mesenchymal stem cell transplantation. Zhonghua Er Ke Za Zhi 2006;44:787-91.
- Kumarasamy S, Gopalakrishnan K, Shafton A, et al. Mitochondrial polymorphisms in rat genetic models of hypertension. Mamm Genome 2010;21:299-306.
- Weiss H, Arndt T, Jörns A, et al. The mutation of the LEW.1AR1-iddm rat maps to the telomeric end of rat chromosome 1. Mamm Genome 2008;19:292-7.
- Pravenec M, Hyakukoku M, Houstek J, et al. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic strains. Genome Res 2007;17:1319-26.
- Le Blanc S, Villarroel P, Candia V, et al. Type 2 diabetic patients and their offspring show altered parameters of iron status, oxidative stress and genes related to mitochondrial activity. Biometals 2012;25:725-35.
- Fendt L, Niederstätter H, Huber G, et al. Accumulation of mutations over the entire mitochondrial genome of breast cancer cells obtained by tissue microdissection. Breast Cancer Res Treat 2011;128:327-36.
- Hung WY, Wu CW, Yin PH, et al. Somatic mutations in mitochondrial genome and their potential roles in the progression of human gastric cancer. Biochim Biophys Acta 2010;1800:264-70.
- Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res 2009;19:802-15.
- Scatena R. Mitochondria and cancer: a growing role in apoptosis, cancer cell metabolism and dedifferentiation. Adv Exp Med Biol 2012;942:287-308.
- Fogg VC, Lanning NJ, Mackeigan JP. Mitochondria in cancer: at the crossroads of life and death. Chin J Cancer 2011;30:526-39.
- Khan SG, Saxena R, Bickers DR, et al. Inhibition of ras p21 membrane localization and modulation of protein kinase C isozyme expression during regression of chemical carcinogen-induced murine skin tumors by lovastatin. Mol Carcinog 1995;12:205-12.
- Bianchi A, Mariani S, Beggiato E, et al. Distribution of T-cell signalling molecules in human myeloma. Br J Haematol 1997;97:815-20.
- Zhu T, Tsuji T, Chen C. Roles of PKC isoforms in the induction of apoptosis elicited by aberrant Ras. Oncogene 2010;29:1050-61.
- Jasinski P, Welsh B, Galvez J, et al. A novel quinoline, MT477: suppresses cell signaling through Ras molecular pathway, inhibits PKC activity, and demonstrates in vivo anti-tumor activity against human carcinoma cell lines. Invest New Drugs 2008;26:223-32.
- Zhu T, Chen L, Du W, et al. Synthetic Lethality Induced by Loss of PKC δ and Mutated Ras. Genes Cancer 2010;1:142-51.
- Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle 2009;8:1688-97.
- Freeman AK, Morrison DK. 14-3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol 2011;22:681-7.
- Jiang F, Qiu Q, Khanna A, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res 2009;7:330-8.
- Muzio G, Maggiora M, Paiuzzi E, et al. Aldehyde dehydrogenases and cell proliferation. Free Radic Biol Med 2012;52:735-46.
- Jelski W, Kutylowska E, Laniewska-Dunaj M, et al. Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) as candidates for tumor markers in patients with pancreatic cancer. J Gastrointestin Liver Dis 2011;20:255-9.
- van den Hoogen C, van der Horst G, Cheung H, et al. The aldehyde dehydrogenase enzyme 7A1 is functionally involved in prostate cancer bone metastasis. Clin Exp Metastasis 2011;28:615-25.
- Kim DH, Bae J, Lee JW, et al. Proteomic analysis of breast cancer tissue reveals upregulation of actin-remodeling proteins and its relevance to cancer invasiveness. Proteomics Clin Appl 2009;3:30-40.
- Chan KT, Creed SJ, Bear JE. Unraveling the enigma: progress towards understanding the coronin family of actin regulators. Trends Cell Biol 2011;21:481-8.
- Wu L, Peng CW, Hou JX, et al. Coronin-1C is a novel biomarker for hepatocellular carcinoma invasive progression identified by proteomics analysis and clinical validation. J Exp Clin Cancer Res 2010;29:17.