Argonaute protein as a linker to command center of physiological processes
Introduction
Small RNA molecules (small RNAs), including microRNAs (miRNAs), small interfering RNA (siRNAs) and Piwi-interacting RNA (piRNAs), play important roles in cellular processes as diverse as development, disease, stress response, or transposon silencing (1,2). In animals, most endogenous miRNAs are transcribed by RNA polymerase II (Pol II) to generate a primary miRNA (pri-miRNA) containing imperfectly base-paired hairpin structures. And then the pri-miRNA is bound to dsRNA binding domain (dsRBD) protein DGCR8, a homologue of Drosophila melanogaster (D. melanogaster) Pasha, forming the microprocessor complex together with Drosha. Then DGCR8 directs Drosha to cleave 11 bp away from the dsRNA-ssRNA junction to release approximately 60-70 nt precursor miRNAs (pre-miRNAs). And later, pre-miRNAs are exported to the cytoplasm by expotin-5 where they are subjected to the second processing. With the help of Dicer, a member of RNase III family, an imperfect pairing about 22 nt miRNA:miRNA* duplex with 2-base pair 3' overhangs at both ends is yielded. In plants, the dsRBD protein is HYPONASTIC LEAVES 1 (HYL1) and there exists no Drosha homologue. And the production of Dicer-like (DCL) is always in nucleus (1,3). The siRNAs synthesis in D. melanogaster also need Dicer (dicer1, dicer2), associating with a dsRBD protein partner, Loqs and R2D2, respectively, whereas the maturation of piRNA needs Piwi or Aubergine (Aub) instead of Dicer protein. These small RNAs mediated gene silencing are involved in regulating diverse biological processes, and Argonaute (AGO) protein is the essential component of the RNA-induced silencing complex (RISC). Only one strand of mature miRNA is selectively incorporated into miRNA-protein complex (miRNP/miRISC) and binds to complementary sites in 3' untranslated regions (3' UTRs) of the target. Imperfect base pairing of the miRNA with the target results in translational inhibition, while perfect base pairing with its transcript promotes RNA cleavage. A new research suggested that the overexpression of AGO protein decelerates miRNA degradation and increases miRNA half-life. In simple terms, AGO proteins can stabilize mature miRNAs (4). As miRNA control has emerged as a critical regulatory principle in the mammalian physiological processes, there is no doubt that AGO proteins participate in molecular regulation of the small RNAs functions. Furthermore, miRNA, siRNA and to piRNA are only known to be present in eukaryotes, but AGO proteins have been reported in various organisms including bacterial and archaea species in recent years. The fact suggests that prokaryotes may use different mechanisms in an attempt to regulate translation of target mRNAs, and to provide protection against viruses and transposable elements. Recently, the significant roles of AGO proteins in maintaining genome integrity, controlling RNA stability and protein synthesis are confirmed. Furthermore, human AGO proteins have been reported to be associated with kinds of diseases, although little is known about their molecular mechanism, for example, colonic carcinogenesis. In the following, we present a review about the evolution, functional structure, subcellular location, and post-translational modification of AGO proteins, and the discussion may help us understand their functions in physiology, human disease and antiviral defense.
Evolution of AGO protein
AGO proteins are widely involved in many cellular physiological processes through RNA silencing pathways, but the total number of members varies across different species, such as 10 AGO proteins in Arabidopsis, 8 in Homo sapiens, 1 in Schizosaccharomyces pombe, etc. Obviously, evolutionary analyses of AGO proteins are conducive to understand function diversification in development and physiology. AGO proteins can be grouped into three major clades in eukaryotes, the Ago clade proteins (Ago), the PIWI clade proteins (PIWI) and worm-specific clade proteins (WAGO) (2,5,6). The Ago loaded with miRNA or siRNA ensures that the appropriate guide strand is selectively stabilized (1), however, piRNA is loaded into PIWI (7). What’s more, Ago extensively exists in all species, including animals, plants and fungi. PIWI is only found in animal’s germ cells and germline stem cells, which participates in the biogenesis and cellular functions of piRNAs in animal gonads and functions in genome surveillance (8). In this paper, 135 AGO protein sequences from 36 species were chosen for phylogenetic analysis (Figure 1A). The results showed that genes encoding AGO proteins have undergone a marked degree of duplication, especially in plants and metazoans, and the champion of this expansion is Caenorhabditis elegans (C. elegans). In C. elegans, the member C06A1.4 has been certificated as pseudogene, and the other 25 AGO proteins were found in different clades, such as Ce_Alg1, Ce_Rde1 in Ago; Ce_Prg1, R09a1.1 in PIWI; and Csr-1, Ppw-1 and the rest in WAGO clade. However, the phylogenetic distribution of archaeal and bacterial AGO proteins is chaotic, and both species consist of a phylogenetic clade away from eukaryotic AGO proteins, suggesting that multiple horizontal gene transfers (HGT) have occurred between bacteria and archaea. The above result is consistent with the notion that prokaryotic AGO (pAGO) genes mostly disseminated by HGT (9).
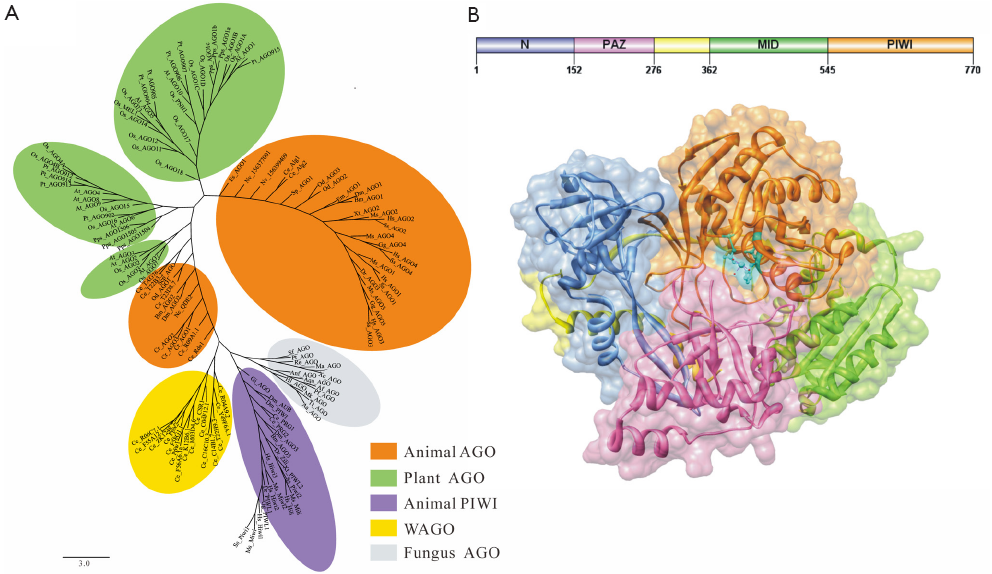
Characteristics of AGO protein structure
AGO proteins are multidomain proteins expressed in all kinds of organisms. As the functions of pAGO protein are still unclear, eukaryotic AGO proteins are taken as excellent models for functional research in prokaryotes. AGO proteins are composed of 4 discrete folding units, which are N-terminal region, PAZ domain, MID domain and PIWI domain, respectively (Figure 1B). The N-terminal region acts as a wedge, interceding in the trajectory of the annealed guide/target duplex (10). A new study shows that a functional N domain is strictly required for small RNA duplex unwinding but not for precedent duplex loading or subsequent target cleavage (11). PAZ domain is approximately 140 aa in length that is identified in both AGO proteins and Dicer enzymes, containing a binding pocket which is used to anchor the 2-nucleotide 3'-overhang of siRNA-like duplex (12,13). Nevertheless, we found no PAZ domain in most prokaryotes by domain scanning, so it led us to suppose that the domain may be not so important in prokaryotes. MID domain, which is located between the PAZ domain and the PIWI domain, is similar to the glucose/galactose-binding protein (GGBP) and Lac repressor (14). The main function of MID domain is to provide a binding pocket for the 5'-phosphate of guide RNA. Besides, a special ring consisting of P523, G524, K525, T526 and P527 in Hs_AGO2 has been certified, and has the ability to recognize the first nucleotide of a miRNA sequence, showing a strong bias to U or A at the 5' position of miRNA (15). Additionally, Kiriakidou et al. (16) found MC sequence in Hs_AGO2, which is similar to translation initiation factor 4E-like (eIF4E-like) mRNA cap-binding motif, and proposed that the sequence may bind to target mRNA disturbing translation initiation. Whereas, Kinch et al. (17) mapped the MC sequence of Hs_AGO2 to MID domain, but did not find the motif, meaning that it’s not easy to explain the mechanism of inhibition of translation initiation. Djuranovic et al. (18) superimposed the MID domain of Dm_Ago1 onto the Rossmann-like fold of the archaeal and eubacterial homologues, and the result suggested that the eukaryotic MID domains take the allosteric regulation similar to the prokaryotes. Furthermore, the crystal structural analysis of Neurospora crassa quelling deficient 2 (QDE-2) MID domain showed that there exists an another ligand-binding site previously unreported (19), suggesting a simple mechanistic explanation for the allosteric effect in eukaryotic AGO proteins, but the precise mechanism of MID domain directly involved in the translational suppression of target mRNA remains to be explored.
PIWI domain, which is unique to the AGO proteins superfamily, adopts a classical RNase H fold, and three residues within the PIWI domain form a catalytic triad. Song et al. (20) showed us the similarity of Pf_AGO and RNase H in the PIWI domain, and revealed that PIWI domain has three highly conserved residues (D558, D628, H745), which form the DDH motif colored by cyan in Figure 1B. Two aspartic acids, D558 and D628, were identified as likely active site residues, while the third residue is variable and might be R or K. The experimental analysis of Thermus thermophilus AGO protein bound to a 5’-phosphorylated 21-base DNA guide strand showed the molecular details of guide-strand recognition and cleavage: the bases 2 to 6 of seed sequence are positioned for matching with the mRNA target on the basic principle of complementary base pairing, and bases 10 and 11 at the cleavage site are locked by two critically positioned arginines (21). Further instructions should come from two putative models for target-RNA recognition mediated by AGO protein. The first is fixed-end model which postulate that both ends of the guide RNA remain stably bound to AGO protein during slicing, limiting the seed region to 2-8 nt of the guide RNA by topological constraint. The second is two-state model, and the distinctive feature is 3' end of the guide strand dissociating from the PAZ domain (22,23). Taken Thermus thermophilus AGO protein as an example, the 3' end of the guide switches on and off PAZ domain during the catalytic cycle, whereas, its 5' end remains fixed (10). However, detailed molecular mechanisms of target RNA recognition and high-affinity guide-target interactions in different species remain unclear.
Subcellular location of AGO protein
Localization studies using immunofluorescence approach and tagged Ago protein showed that in somatic cells, Ago protein concentrated in cytoplasm and cytoplasmic processing body (P body, also referred to as GW or Dcp body), known as site of RNA degradation and transient storage of translationally repressed mRNA (24-27). It has been shown that when double-stranded siRNAs are transfected into cells, they rapidly localize to P-bodies, and up-regulate expression of Human_AGO2 and GW182 (28). However, Ago proteins can inhibit translation of target mRNA in the absence of P-bodies. What’s more, a quantitative analysis has revealed that the Ago proteins are not only concentrated in P-bodies but also in stress granules (SGs) (29). It was noted that there are non-translating mRNAs, translation initiation factors (eIF2, eIF3, eIF4E, and eIF4G) and 40S ribosome in SGs, when cells are exposed to a number of stresses including heat shock, osmotic stress and oxidative stress (29-31). Further evidence showed that it is in a miRNA-dependent way that Ago proteins localize to SGs, and physical association between SGs and P-bodies in vivo is promoted by the related mRNA decay factors TTP and BRF1 (32,33).
Nuclear AGOs function in a wide range of biological processes, such as transportation of specific sRNAs to distinct compartments by specific AGO, nuclear RNAi, and ribonucleoprotein (RNP) complex and DNA methylation complex assembly, etc. In the absence of siRNAs, C. elegans Ago NRDE-3 resides in the cytoplasm, otherwise NRDE-3 binds siRNAs redistributes to the nucleus (34). NRDE-3 associates with siRNAs, acting in conjunction with the nuclear RNAi factors NRDE-1, NRDE-2, and NRDE-4, and promotes siRNA expression in inheriting progeny (35). Analysis of retrovirus-infected cells further demonstrated that Ago1 co-localized with siRNA in the nucleus, while Ago2 co-localized with siRNA in the inner nuclear envelope, indicating nuclear compartment distribution of Ago-associated siRNA and a nuclear trafficking mechanism for components of RNA-induced transcriptional gene silencing-like complex involving the actin cytoskeleton (36). A novel nuclear miRNA, miR-671, guides Ago2 to specifically cleave the antisense. Blocking miR-671 led to an upregulation of circular cerebellar degeneration-related protein 1 (CDR1) antisense and sense mRNA levels in HEK293 cells. And in the presence of miR-671-Ago2-RISC in the nucleus, the circular CDR1 non-coding antisense is degraded, destabilizing the sense mRNA. This may be a novel mechanism for gene regulation that involves stabilization of a sense transcript by a circular non-coding antisense RNA (37). Interestingly, Ago proteins are also found in the Cajal bodies (CBs), originally called them nucleolar “accessory” bodies (38,39), known as site of siRNA and miRNA biogenesis in plants, whereas in animals siRNA and miRNA are finally produced in cytoplasm. Li et al. (40) reported that siRNA processing center shares features of CBs, and the DCL3/AGO4-containing nuclear processing center co-localize with CBs in Arabidopsis thaliana. Also, CBs act as the key site of nuclear RNP complex assembly, and it has been suggested that a larger DNA methylation complex contains At_AGO4 and siRNAs (40,41). In short, AGO can regulate nuclear events like transcription, genome maintenance, and splicing (42).
In contrast, PIWI family has not been shown to interact with miRNAs and does not localize to cytoplasmic foci when ectopical expression of Hiwi (24). There are three members of PIWI proteins in D. melanogaster: Aub, AGO3 and Piwi [a polar granule (PG) component]. It has been proven by confocal immunofluorescence and immuno-electron microscopy methods that all of them are required for germ cell formation and germline stem cell maintenance (43,44). The PG is a perinuclear germline structure, most notably during oogenesis as the nuage in flies, and the counterpart of PG in mammalian is chromatoid body, which is a male reproductive cell-specific organelle (45). Furthermore, chromatoid bodies are related to the P Bodies, which concentrate the enzymes involved in the RNA-decay pathway and accumulate mRNAs targeted for degradation or translation repression (25,45,46), and seem to operate as an intracellular nerve-center of the miRNA pathway. Surprisingly, Mus musculus Piwi (Miwi), which is essential for spermatogenesis, is found in RNP complex in male germ cells, and is co-localized with other Ago/PIWI members, such as AGO2 and AGO3 in the chromatoid body (45).
Posttranslational modification of AGO protein
sRNAs and their associated proteins are subject to diverse modifications contributing to the dynamic regulation of RNA silencing, and post-translational modifications (PTMs) of RNA silencing proteins include phosphorylation, hydroxylation, ubiquitination, methylation, etc. The first genetic demonstration showed that only Hs_AGO2 (known as eIF2C2) has endonucleolytic activity, although the postulated catalytic residues (Asp587, Asp669 and His807) of the DDH motif are conserved in other human AGO proteins. Now a major breakthrough has been achieved in the study of PTMs of Hs_AGO2 (Figure 2A) (47-50).
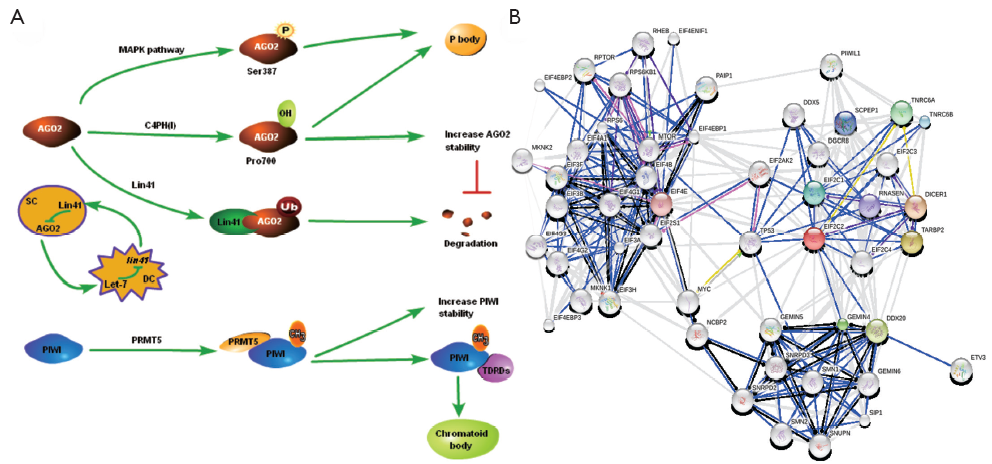
Using in vivo labeling and mass spectrometry (MS) approach, Zeng et al. (47) found that Hs_AGO2 can be phosphorylated at Ser387, which is significantly induced by treatment with sodium arsenite, a known activator of p38 mitogen-activated protein kinase (p38 MAPK) signaling pathway. What’s more, the p38 MAPKs are a second mammalian stress-activated MAPK family. Further experiments showed that Hs_AGO2 is phosphorylated at the same site directly by MAPKAPK2 in vitro. Anisomycin, another p38 activator, also induces the phosphorylation of Hs_AGO2 at Ser-387, indicating that p38 MAPK activation via different mechanisms is sufficient to induce Hs_AGO2 phosphorylation. The results show that Hs_AGO2-mediated gene silencing links to the MAPK signaling pathways, or rather, Hs_AGO2 could be activated by a wide variety of different stress stimuli acting through the p38 MAPK pathway. In fact, mutating Ser (387) to alanine (GRA or ARG) leads to significantly reduced localization of Hs_AGO2 to cytoplasmic P-bodies. It is notable that the recent emerging evidence shows that p38 stress MAPK pathway may function as a tumor suppressor through regulating Ras-dependent and -independent proliferation, transformation, invasion and cell death by isoform-specific mechanisms.
In proteomic analysis of human AGO proteins, Qi et al. (48) reported that Hs_AGO2 physically interacts with the a-[P4H-a(I)] and b-(P4H-b) subunits of the type I collagen prolyl-4-hydroxylase [C-P4H(I)], which catalyzes the formation of the 4-hydroxyproline residues that are critical for the generation of triple helical collagen molecules. Hydroxylation of the Hs_AGO2 at Pro700 has been experimentally identified by mass spectrometric analysis in living cell, and both Hs_AGO2 and Hs_AGO4 seem to be more efficiently hydroxylated than Hs_AGO1 and Hs_AGO3 by recombinant human C-P4H(I). Conversely, knockdown of C-P4H(I) reduces siRNA-induced silencing activity in vitro. Mutation of the Pro700 to GRA (P700A) results in less stable than the wild-type Hs_AGO2, but has no effect on Hs_AGO1 and Hs_AGO3, suggesting that Pro700 is important for Hs_AGO2 in stability regulation (48). Obviously, post-translational hydroxylation of protein is not only required for the stability of Hs_AGO2, but also has been proven to involved in the localization to the P-bodies.
Apart from phosphorylation and hydroxylation, ubiquitination also plays an important role in regulating Hs_AGO2 stability. The first lin-4 miRNA was discovered by Lee et al. (51), however, it didn’t give rise to a lot of attention until let-7 miRNA had been found (52). And then it is also found in plants, mouse and human (53). The target genes of let-7 are lin-41, lin-42, lin-14, lin-28, etc. Lin-41 is among the best studied regulatory targets of let-7 in C. elegans (54), and it encodes a conserved protein containing a tripartite motif (TRIM), which frequently possesses E3 ubiquitin ligase activity. Also, mouse homologue of lin-41 (mLin-41) exhibits an E3 ubiquitin ligase activity in an auto-ubiquitylation assay, and mediated ubiquitylation of Ms_AGO2 in vitro and in vivo (55). In addition, Lin-41 is negatively regulated by let-7, and let-7 is highly expressed in differentiated cell (DC) but not in the stem cell (SC). Consequently, the expression of Lin-41 is higher in SC than in DC. Thereby, mLin-41 and let-7 constitute a negative feed-back circuit shown in Figure 2A.
Most animal PIWI contain symmetric arginine dimethylation (sDMA) motifs composed of an arginine flanked by either two glycines (GRG) or by a GRG and an GRA (56,57), and sDMA modifications are typically clustered close to the amino terminus, but no animal Ago containing such motifs. Further evidence of PIWI dimethylation modification came occasionally that PIWI protein couldn’t be methylated in dPRMT5 deficient ovaries, showing that methyltransferase PRMT5 catalyzes sDMAs of Piwi, AGO3 and Aub in flies ovaries in vivo (49,58). As arginine methylation helps to stabilize PIWI and consequently increases piRNA levels, leading to transposon silencing and germ cell development. sDMA modification of Aub is required for interaction with Tudor domain-containing proteins (TDRDs) in D. melanogaster (59,60). It is notable that Tudor associates with PIWI protein specifically in gametogenesis (61,62), and contributes to Aub localization to the posterior pole of the embryo (63). As sDMA is a covalent marking of proteins by methyl group addition to specific arginine residues (64,65), the precise positions of sDMAs in mouse and flies PIWI are identified using the MS method, and the results showed that there are three modified arginines in Miwi (R14, R49, and R371) and six in Mili (R74, R95, R100, R146, R163, and R549), respectively (59). In addition, R11, R13 and R15 in Aub and R4, R68 and R70 in AGO3 were determined to be sDMAs in flies (66). An intriguing finding is that methylated PIWI can interact with the TDRDs and, then, locate to chromatoid body in mouse. There are different opinions arguing that the stability of PIWI is not changed by the loss of sDMA modification in D. melanogaster germline.
AGO protein interaction with other proteins
Recently, proteomic and functional analyses of Human AGO proteins revealed that they play crucial roles in the protein regulatory network governing the growth and development of an organism (67). Hs_AGO2 (EIF2C2) has endonuclease activity described previously, and interacts with Dicer1 and TAR (HIV-1) RNA binding protein 2 (TARBP2) which recruits the Dicer1 complex to Hs_AGO2 for miRNA processing and gene silencing (68). DDX20 (GEMIN3), GEMIN4, TNRC6A, EIF2C1 and TNRC6B interact with Hs_AGO2, and these proteins are required for miRNA-dependent translational repression or endonucleolytic cleavage of complementary mRNAs (69-71). Also, AGO2 interacts with TP53 and MYC and so on (Figure 2B). AGO2 interacts with the retroviral Group Specific Antigen (GAG) core proteins and preferentially binds unspliced RNAs through the RNA packaging sequences without affecting RNA stability or eliciting translation repression (72). While a recent study indicated that major components of the miRNA pathway, such as DGCR8, AGO1, AGO2, PACT, and TARBP1, are dysregulated in epithelial skin cancer (73). Transmembrane human prion protein PrPC preferentially binds AGO, and PrPC promotes formation or stability of miRISC effector complexes and miRNA-repressed mRNA (74). As protein-protein interactions form the basis for a vast majority of cellular events, including signal transduction and transcriptional regulation, the above results suggest an important associoation between Hs_AGO2 and many human diseases, such as autoimmune diseases (75,76), Fragile X syndrome (77,78), tumor (79) and antiviral defence (80). The anti-Su antibodies from both human patients with rheumatic diseases and a mouse model of autoimmunity recognize the endonucleolytic AGO and Dicer proteins (75,76), and a later study declared that autoantibodies to the Su antigen are found in a variety of systemic rheumatic diseases, suggesting that AGO proteins may involve in autoimmune diseases. Whereas Fragile X syndrome is caused by loss of expression of the fragile X mental retardation protein (FMRP), and it has been reported that mammalian FMRP interacts with miRNAs and AGO2. Overexpression of dFmr1 gene could lead to the partial loss of AGO1 and suppress the neuronal apoptosis, showing that AGO1 is critical for FMRP function in neural development and synaptogenesis. Wherase AGO2 together with FXR1P, one of two autosomal paralogs of the FMRP, factors associated with miRNPs, are recruited by AU-rich elements (AREs) in 3’ UTR of cytokine mRNAs, such as tumor necrosis factor-α (TNFα), contributing significantly to translation up-regulation upon cell cycle arrest (78,81,82). Li et al. (79) identified that AGO1-4 and PIWIL1-4 are significantly higher in tumorous tissue than in adjacent tissue, indicating that they should be capable of promoting tumor invasion. Similarly, high expression of Hili can also contribute to tumorigenesis (83). Also, increased mRNA levels of Hiwi, which has been described primarily in hematopoietic stem cell (HSC) and germ cells, have been detected in several distinct types of tumors, such as seminoma cell hyperplasia, adenocarcinoma of the pancreas (84), esophageal squamous cell carcinoma (85) and soft-tissue sarcoma (86). miR-122 is liver-specific miRNA for hepatitis C virus (HCV), binding the 5' UTR of the messenger-sense HCV RNA genome, stimulating translation and promoting genome replication, and binding HCV RNA in association with Ago2 involved in slowing decay of the viral genome in infected cells. Namely, Ago2 and miR-122 act coordinately to protect the viral genome from 5' exonuclease activity of the host mRNA decay machinery (87). More AGO protein-mediated miRNA functions involved in physiological and pathophysiological processes are shown in Table 1. Do they integrate with their associated sRNAs to prompt tumorigenesis by RNA silence way, or interact with other proteins resulting in tumor? Co-expression of Ago and shRNA showed that Hs_AGO2 is the primary rate-limiting determinant of both in vitro and in vivo RNAi efficacy (106). Another interesting evidence is that in Arabidopsis both AGO1 and AGO2 play crucial roles in antiviral defense (80), and this may help further understanding of functional differentiation in AGO proteins.
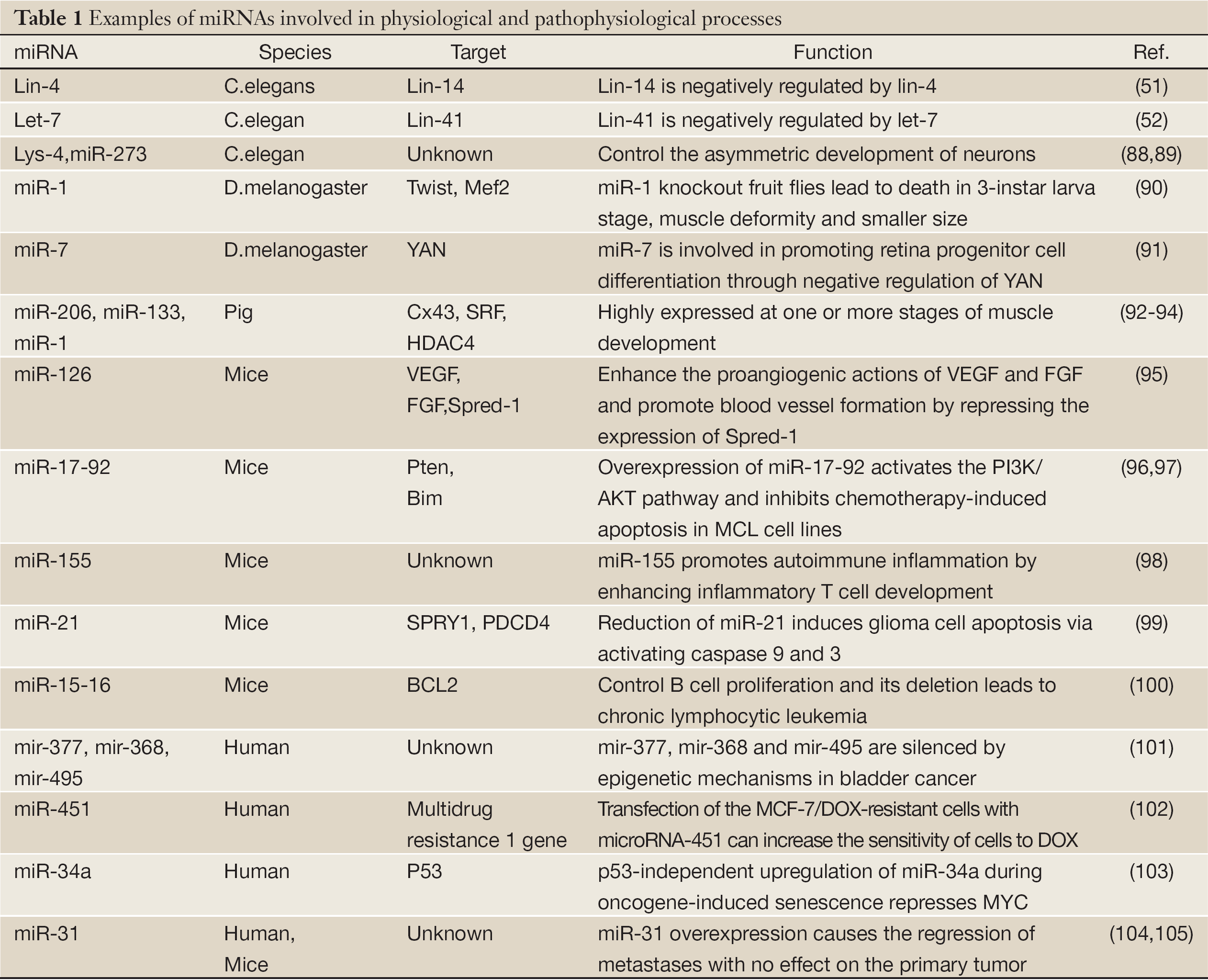
Full Table
Conclusions
AGO proteins are key components of small RNA pathways and are involved in diverse biological functions and pathogenesis of diseases. It has got dramatic advances in AGO structure and regulation of its function. As discussed earlier, overexpression of miR-17-92 may activate the PI3K/AKT pathway and inhibit chemotherapy-induced apoptosis in MCL cell lines, and miR-17-92 has been considered as a therapeutic target. In fact, AGO proteins in conjunction with the different small regulatory RNAs are associated with various cellular processes, such as stem cell self-renewal, tumor development and progression, and so on. Only with the help of AGO proteins, human physiological processes can run smoothly to keep your body healthy. The elucidation of biochemical and physiological functions of AGO proteins will facilitate the determination of the diagnostic and/or prognostic value of AGO proteins. Also, AGO proteins may serve as potential biomarkers in early detection of human diseases. Despite so much progresses having been made, the identification of molecular basis of physiological processes and pathology still is a challenging problem. The more detailed analysis of AGO protein functions may help us deeply understand the biology of disease and facilitate development of drugs to slow, stop or reverse disease progression.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 2011;12:19-31. [PubMed]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008;9:22-32. [PubMed]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009;136:642-55. [PubMed]
- Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol 2011;8:1149-57. [PubMed]
- Yigit E, Batista PJ, Bei Y, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 2006;127:747-57. [PubMed]
- Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol 2007;3:36-43. [PubMed]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008;135:1201-14. [PubMed]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126-39. [PubMed]
- Makarova KS, Wolf YI, van der Oost J, et al. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct 2009;4:29. [PubMed]
- Parker JS. How to slice: snapshots of Argonaute in action. Silence 2010;1:3. [PubMed]
- Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol 2012;19:145-51. [PubMed]
- Lingel A, Simon B, Izaurralde E, et al. Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol 2004;11:576-7. [PubMed]
- Yuan YR, Pei Y, Ma JB, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 2005;19:405-19. [PubMed]
- Hall TM. Structure and function of argonaute proteins. Structure 2005;13:1403-8. [PubMed]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010;465:818-22. [PubMed]
- Kiriakidou M, Tan GS, Lamprinaki S, et al. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 2007;129:1141-51. [PubMed]
- Kinch LN, Grishin NV. The human Ago2 MC region does not contain an eIF4E-like mRNA cap binding motif. Biol Direct 2009;4:2. [PubMed]
- Djuranovic S, Zinchenko MK, Hur JK, et al. Allosteric regulation of Argonaute proteins by miRNAs. Nat Struct Mol Biol 2010;17:144-50. [PubMed]
- Boland A, Tritschler F, Heimstädt S, et al. Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep 2010;11:522-7. [PubMed]
- Song JJ, Smith SK, Hannon GJ, et al. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004;305:1434-7. [PubMed]
- Wang Y, Sheng G, Juranek S, et al. Structure of the guide-strand-containing argonaute silencing complex. Nature 2008;456:209-13. [PubMed]
- Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell 2005;122:17-20. [PubMed]
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature 2009;457:405-12. [PubMed]
- Liu J, Valencia-Sanchez MA, Hannon GJ, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 2005;7:719-23. [PubMed]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 2007;8:9-22. [PubMed]
- Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol 2005;7:633-6. [PubMed]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell 2007;25:635-46. [PubMed]
- Jagannath A, Wood MJ. Localization of double-stranded small interfering RNA to cytoplasmic processing bodies is Ago2 dependent and results in up-regulation of GW182 and Argonaute-2. Mol Biol Cell 2009;20:521-9. [PubMed]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A 2006;103:18125-30. [PubMed]
- Anderson P, Kedersha N. RNA granules. J Cell Biol 2006;172:803-8. [PubMed]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol 2009;21:403-8. [PubMed]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009;36:932-41. [PubMed]
- Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 2005;169:871-84. [PubMed]
- Guang S, Bochner AF, Pavelec DM, et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 2008;321:537-41. [PubMed]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2011;108:19683-8. [PubMed]
- Ahlenstiel CL, Lim HG, Cooper DA, et al. Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Res 2012;40:1579-95. [PubMed]
- Rossi JJ. A novel nuclear miRNA mediated modulation of a non-coding antisense RNA and its cognate sense coding mRNA. EMBO J 2011;30:4340-1. [PubMed]
- Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev 2008;18:197-203. [PubMed]
- Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol 2005;21:105-31. [PubMed]
- Li CF, Pontes O, El-Shami M, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 2006;126:93-106. [PubMed]
- Höck J, Meister G. The Argonaute protein family. Genome Biol 2008;9:210. [PubMed]
- Gagnon KT, Corey DR. Argonaute and the nuclear RNAs: new pathways for RNA-mediated control of gene expression. Nucleic Acid Ther 2012;22:3-16. [PubMed]
- Megosh HB, Cox DN, Campbell C, et al. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol 2006;16:1884-94. [PubMed]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 2009;25:355-76. [PubMed]
- Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol 2007;8:85-90. [PubMed]
- Kotaja N, Bhattacharyya SN, Jaskiewicz L, et al. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A 2006;103:2647-52. [PubMed]
- Zeng Y, Sankala H, Zhang X, et al. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J 2008;413:429-36. [PubMed]
- Qi HH, Ongusaha PP, Myllyharju J, et al. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 2008;455:421-4. [PubMed]
- Heo I, Kim VN. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell 2009;139:28-31. [PubMed]
- Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell 2010;143:703-9. [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [PubMed]
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901-6. [PubMed]
- Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000;408:86-9. [PubMed]
- Slack FJ, Basson M, Liu Z, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 2000;5:659-69. [PubMed]
- Rybak A, Fuchs H, Hadian K, et al. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 2009;11:1411-20. [PubMed]
- Krause CD, Yang ZH, Kim YS, et al. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther 2007;113:50-87. [PubMed]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell 2005;18:263-72. [PubMed]
- Kirino Y, Kim N, de Planell-Saguer M, et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 2009;11:652-8. [PubMed]
- Vagin VV, Wohlschlegel J, Qu J, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 2009;23:1749-62. [PubMed]
- Liu H, Wang JY, Huang Y, et al. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev 2010;24:1876-81. [PubMed]
- Vourekas A, Kirino Y, Mourelatos Z. Elective affinities: a Tudor-Aubergine tale of germline partnership. Genes Dev 2010;24:1963-6. [PubMed]
- Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev 2010;24:636-46. [PubMed]
- Kirino Y, Vourekas A, Sayed N, et al. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 2010;16:70-8. [PubMed]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell 2009;33:1-13. [PubMed]
- Wolf SS. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci 2009;66:2109-21. [PubMed]
- Nishida KM, Okada TN, Kawamura T, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 2009;28:3820-31. [PubMed]
- Höck J, Weinmann L, Ender C, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 2007;8:1052-60. [PubMed]
- Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005;436:740-4. [PubMed]
- Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP: mRNA association in polyribosomes in a human neuronal cell line. RNA 2004;10:387-94. [PubMed]
- Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 2009;15:1059-66. [PubMed]
- Perron MP, Provost P. Protein interactions and complexes in human microRNA biogenesis and function. Front Biosci 2008;13:2537-47. [PubMed]
- Bouttier M, Saumet A, Peter M, et al. Retroviral GAG proteins recruit AGO2 on viral RNAs without affecting RNA accumulation and translation. Nucleic Acids Res 2012;40:775-86. [PubMed]
- Sand M, Skrygan M, Georgas D, et al. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol Carcinog 2012;51:916-22. [PubMed]
- Gibbings D, Leblanc P, Jay F, et al. Human prion protein binds Argonaute and promotes accumulation of microRNA effector complexes. Nat Struct Mol Biol 2012;19:517-24, S1.
- Jakymiw A, Ikeda K, Fritzler MJ, et al. Autoimmune targeting of key components of RNA interference. Arthritis Res Ther 2006;8:R87. [PubMed]
- Pruijn GJ. The RNA interference pathway: a new target for autoimmunity. Arthritis Res Ther 2006;8:110. [PubMed]
- Jin P, Zarnescu DC, Ceman S, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci 2004;7:113-7. [PubMed]
- Li Y, Lin L, Jin P. The microRNA pathway and fragile X mental retardation protein. Biochim Biophys Acta 2008;1779:702-5.
- Li L, Yu C, Gao H, et al. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer 2010;10:38. [PubMed]
- Harvey JJ, Lewsey MG, Patel K, et al. An antiviral defense role of AGO2 in plants. PLoS One 2011;6:e14639. [PubMed]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007;318:1931-4. [PubMed]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 2007;128:1105-18. [PubMed]
- Ye Y, Yin DT, Chen L, et al. Identification of Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS One 2010;5:e13406. [PubMed]
- Grochola LF, Greither T, Taubert H, et al. The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer 2008;99:1083-8. [PubMed]
- He W, Wang Z, Wang Q, et al. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer 2009;9:426. [PubMed]
- Taubert H, Greither T, Kaushal D, et al. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene 2007;26:1098-100. [PubMed]
- Shimakami T, Yamane D, Jangra RK, et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A 2012;109:941-6. [PubMed]
- Chang S, Johnston RJ Jr, Frøkjaer-Jensen C, et al. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 2004;430:785-9. [PubMed]
- Miska EA, Alvarez-Saavedra E, Abbott AL, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 2007;3:e215. [PubMed]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev 2005;19:2343-54. [PubMed]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 2005;123:1267-77. [PubMed]
- Huang TH, Zhu MJ, Li XY, et al. Discovery of porcine microRNAs and profiling from skeletal muscle tissues during development. PLoS One 2008;3:e3225. [PubMed]
- McDaneld TG, Smith TP, Doumit ME, et al. MicroRNA transcriptome profiles during swine skeletal muscle development. BMC Genomics 2009;10:77. [PubMed]
- Liu HC, Hicks JA, Trakooljul N, et al. Current knowledge of microRNA characterization in agricultural animals. Anim Genet 2010;41:225-31. [PubMed]
- Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 2008;15:261-71. [PubMed]
- Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 2008;9:405-14. [PubMed]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell 2009;136:26-36. [PubMed]
- O’Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010;33:607-19. [PubMed]
- Ma X, Kumar M, Choudhury SN, et al. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc Natl Acad Sci U S A 2011;108:10144-9. [PubMed]
- Klein U, Lia M, Crespo M, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010;17:28-40. [PubMed]
- Zhang L, Volinia S, Bonome T, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A 2008;105:7004-9. [PubMed]
- Kovalchuk O, Filkowski J, Meservy J, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 2008;7:2152-9. [PubMed]
- Christoffersen NR, Shalgi R, Frankel LB, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ 2010;17:236-45. [PubMed]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 2011;11:849-64. [PubMed]
- Valastyan S, Chang A, Benaich N, et al. Activation of miR-31 function in already-established metastases elicits metastatic regression. Genes Dev 2011;25:646-59. [PubMed]
- Grimm D, Wang L, Lee JS, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest 2010;120:3106-19. [PubMed]
