Effect of cytotoxic T-lymphocyte antigen-4, TNF-alpha polymorphisms on osteosarcoma: evidences from a meta-analysis
Introduction
Osteosarcoma is the most common malignancy of bone and has peak occurrence in children and young adults (1). The exact mechanisms of osteosarcoma are largely unknown, but many evidences have suggested that multiple genetic and environmental factors may play important roles in the carcinogenesis of osteosarcoma. Many studies have confirmed that the T cell response plays an important role in the immune system, including the antitumor effect, therefore, genes encoding T cell response molecules may potentially affect the carcinogenesis of cancers (2). Cytotoxic T-lymphocyte antigen-4 (CTLA-4) is a costimulatory molecule; it is expressed by activated T cells and is able to down-regulate T-cell proliferation and activation (3,4). Previously, several studies identified that CTLA-4 is able to affect T cell responses in animal tumor models and cancer immunotherapy trials in humans (5-8). Recently, Contardi et al. (9) showed that CTLA-4 molecule is expressed and functions on human tumor cells, such as osteosarcoma and breast cancer, which indicates that CTLA-4 may affect these cancers through multiple pathways. CTLA-4 gene is located on chromosome 2q33 and consists of four exons that encode separate functional domains. Several variants of CTLA-4 gene, such as rs5742909 (–318C/T), rs231775 (+49G/A), and rs3087243 (CT60A/G), are reported to be associated with various cancers (10-12). However, little is known about the association between CTLA-4 polymorphisms and osteosarcoma risk, and the current results about this relationship remained controversial (13-16).
Tumor necrosis factor-alpha (TNF-α) is a cytokine produced by macrophages and monocytes with a wide range of activities, and the polymorphisms of this gene have been found to contribute to major histocompatibility complex (MHC) associations with some autoimmune diseases (17). There are evidences showed that the polymorphisms of TNF-α promoter related to the development and poor prognosis of some cancers, such as gastric cancer (18), prostate cancer (19) and cervical cancer (20). TNF-α rs1800629 (308G/A) polymorphism was wildly studied and there is a functional study suggesting that the A allele of rs1800629 is associated with increased TNF-α production (21). Currently, fewer studies (22-24) investigated the susceptibility of TNF-α polymorphisms to osteosarcoma risk, and their results were inconclusive due to the limited number of subjects.
Recently, there are studies reported that CTLA-4 and TNF-α polymorphisms as a susceptibility marker for recurrent miscarriage (25), and GG genotype of CTLA-4 rs231775 was associated to lower TNF-α levels in patients with chronic hepatitis B virus (HBV) infection (26). Considering the important role of CTLA-4 and TNF-α polymorphisms in the response to cancer carcinogenesis, as well as the inconsistent results of current studies on the association with osteosarcoma risk, we systematically analyzed the CTLA-4 and TNF-α polymorphisms on carcinogenesis of osteosarcoma through a meta-analysis.
Materials and methods
Literature search and data extraction
All methods were based on the guidelines proposed by the Human Genome Epidemiology Network for systematic review of genetic-association studies and followed preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines (27). A systematic literature search was performed on PubMed, EMbase, Cochrane Library, Google Scholar databases, Chinese National Knowledge Infrastructure (CNKI) and conference literature in humans published prior to March 2013 without language restrictions. The search terms included “Osteosarcoma”, “Polymorphism”, “CTLA-4”, “Tumor Necrosis Factor-α” as well as their combinations. We also identified additional studies by screening reference lists of key studies and reviews.
Selection criteria of an eligible study included: (I) studies that investigated the association between the CTLA-4 or TNF-α polymorphism and risk of osteosarcoma; (II) case-control design; (III) studies that reported crude odds ratio (OR) with 95% confidence interval (95% CI) values or sufficient data to calculate crude OR and 95% CI; (IV) for studies with over one publication describing results among the same or overlapping groups of subjects or controls, only the largest of the available published data sets was included; and (V) in order to achieve enough statistical power, we only selected single nucleotide polymorphisms (SNPs) which had been reported by more than two publications.
The following data of the studies were extracted: first author, published year, original country, number of case and control, and genotypes of CTLA-4 and TNF-α. Two investigators (Liu JW and Wang JL) independently extracted the data. The discrepancy in data extraction was resolved by repeating the study review and discussion.
Statistical analysis
Association between CTLA-4 or TNF-α polymorphism and osteosarcoma risk was calculated using crude OR with 95% CI. Four models of allele contrast model, codominant model, dominant model, and recessive model were analyzed respectively. If the P value of the heterogeneity test was >0.05, a fixed-effect model was performed to calculate the combined OR, which assumed the same homogeneity of effect size across all studies. If the P value of the heterogeneity test was <0.05, it showed that the between-study heterogeneity was statistically significant. A random-effects mode was used to calculate the combined OR. Hardy-Weinberg equilibrium (HWE) was calculated in control population to evaluate the quality of the data by using chi-square test. Publication bias was tested graphically by using funnels plots, and the funnel plot asymmetry was assessed by the method of Egger’s test. The statistical analyses were performed using Stata Statistical package (version 11.2; Stata Corp., College Station, TX). All P values were two-sided.
Results
Literature search process
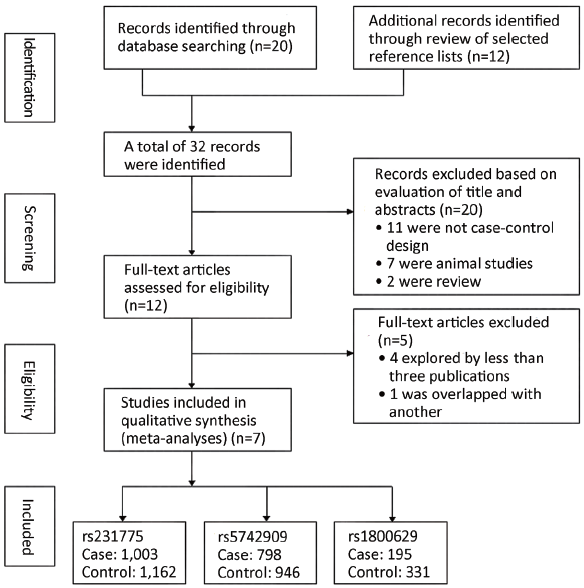
The initial search yielded 32 studies. Twelve potential relevant studies were identified through screening the titles and abstracts. Next by screening the full texts, five studies were excluded. Therefore, a total of seven case-control studies (13-16,22-24) analyzing two SNPs of CTLA-4 (rs231775: +49G/A, rs5742909: –318C/T) and one SNP of TNF-α (rs1800629: –308G/A) were finally included in the present meta-analysis. Figure 1 provides a flow summary of selection process. Table 1 presents the characteristics of individual studies in the meta-analysis and the HWE of controls.
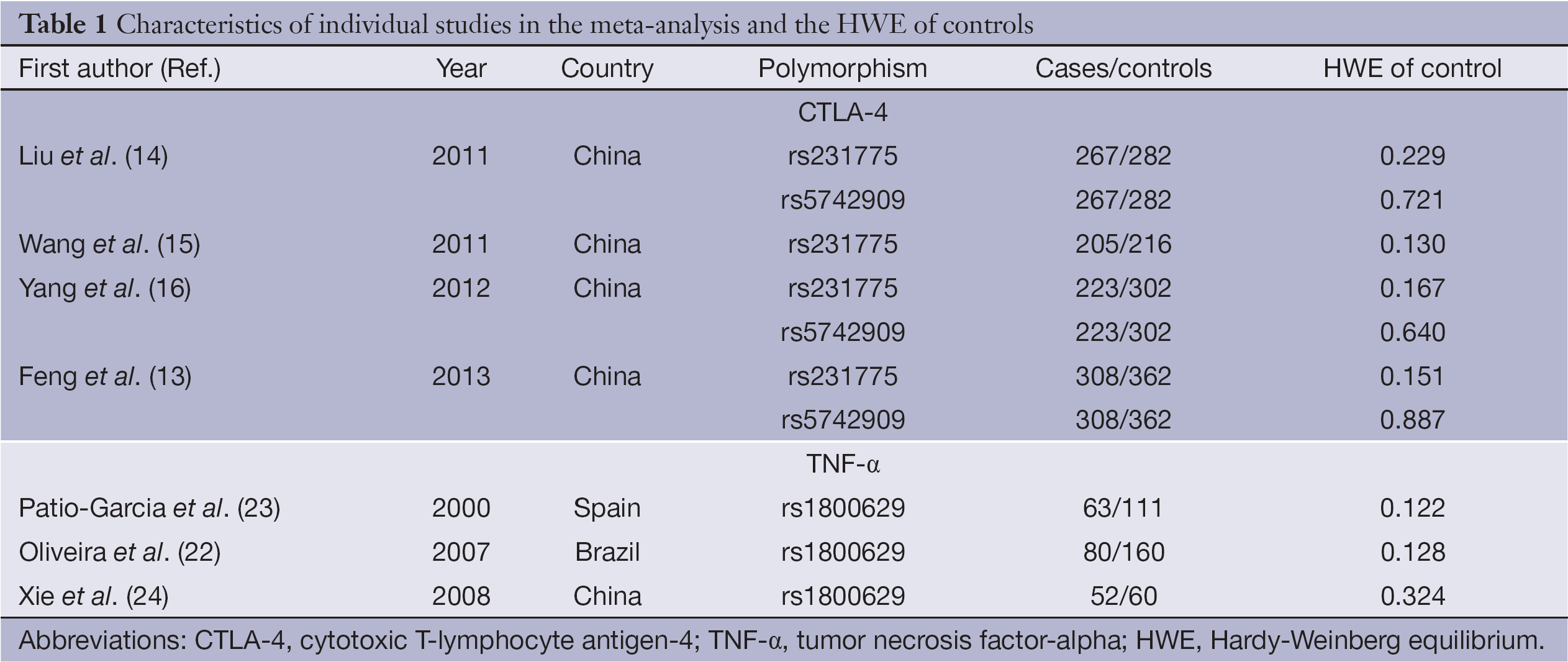
Full table
Quantitative data synthesis
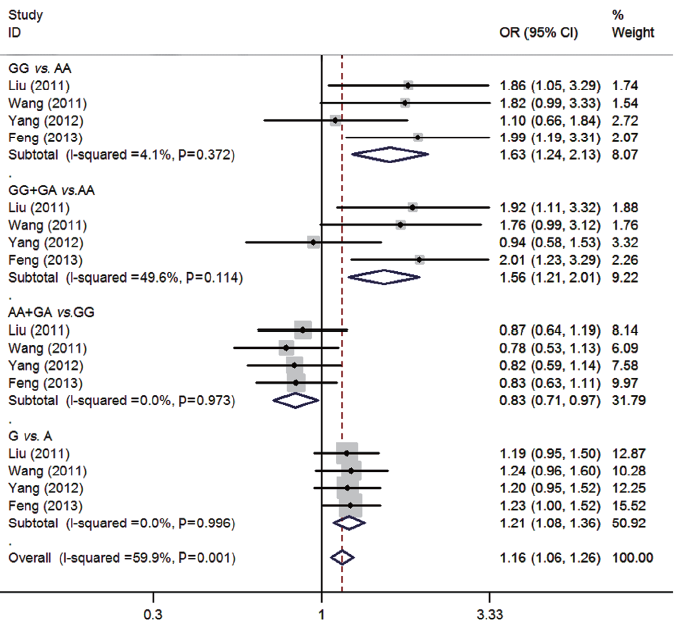
For rs231775 polymorphism of CTLA-4, 4 studies (13-16) consisting of 1,003 osteosarcoma patients and 1,162 controls were synthesized. All the included subjects were Chinese. There were significant associations between rs231775 polymorphism and osteosarcoma risk (GG vs. AA: OR=1.63, 95% CI=1.24-2.13; GG + GA vs. AA: OR=1.56, 95% CI=1.21-2.01; AA + GA vs. GG: OR=0.83, 95% CI=0.71-0.97; G vs. A: OR=1.21, 95% CI=1.08-1.36). No significant heterogeneity was found between studies by Q-test and I2 test (all P>0.05) (Figure 2).
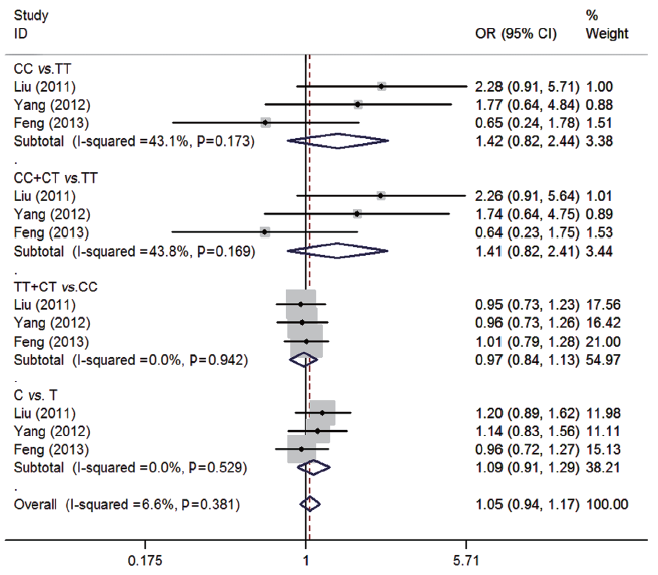
For rs5742909 polymorphism of CTLA-4, three studies (13,14,16) consisting of 798 osteosarcoma patients and 946 controls were synthesized. No significant associations were found between rs5742909 polymorphism and osteosarcoma risk (CC vs. TT: OR=1.42, 95% CI=0.82-2.44; CC + CT vs. TT: OR=1.41, 95% CI=0.82-2.41; TT + CT vs. CC: OR=0.97, 95% CI=0.84-1.13; C vs. T: OR=1.09, 95% CI=0.91-1.29). No between-study heterogeneity was observed (all P>0.05) (Figure 3).
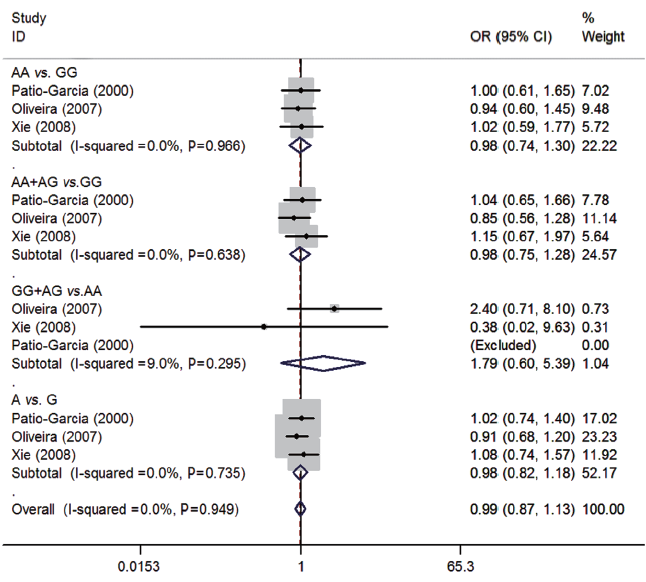
For rs1800629 polymorphism of TNF-α, three studies (22-24) consisting of 195 osteosarcoma patients and 331 controls were synthesized. No significant associations were presented between rs5742909 polymorphism and osteosarcoma risk (AA vs. GG: OR=0.98, 95% CI=0.74-1.30; AA + AG vs. GG: OR=0.98, 95% CI=0.75-1.28; GG + AG vs. AA: OR=1.79, 95% CI=0.60-5.39; A vs. G: OR=0.98, 95% CI=0.82-1.18). No between-study heterogeneity was observed (all P>0.05) (Figure 4).
Publication bias
Genotypes of rs231775, rs5742909 and rs1800629 polymorphisms were plotted against the precision ones using a funnel plot. The result was approximately symmetrical, and no publication bias was observed by Egger’s test (P=0.567 for rs231775 GG + GA vs. AA; P=0.424 for rs5742909 CC + CT vs. TT; P=1.000 for rs1800629 AA + AG vs. GG).
Discussion
CTLA-4 is expressed by cytotoxic T-lymphocytes and binds to B7.1/B7.2 ligands on the surface of antigen-presenting cells, which functions on cell cycle arrest and directly down-regulates T-cell proliferation (3,6,8). CTLA-4 is also known to be able to cause tumor tolerance due to its role in diminishing antitumor responses (9). More and more evidences have shown that CTLA-4 may be correlated with the pathnogenesis of bone diseases, such as rheumatoid arthritis (RA), osteosarcoma, and Ewing’s sarcoma (14,15). CTLA-4 gene consists of four exons that encode separate functional domains: leader sequence, extracellular domain, transmembrane domain, and cytoplasmic domain (2,28).
There are more than one hundred SNPs have been identified in the CTLA-4 gene region (28), in which the rs231775 mutation has been reported as one of the most important polymorphisms. The rs231775 polymorphism is located at position +49 in exon 1, and causes an amino acid exchange (alanine to threonine) in the peptide leader sequence. The A allele of rs231775 at this position is found to be correlated with high expression of CTLA-4 protein (2). Previous studies have evaluated the relationship between this polymorphism and the susceptibility to various autoimmune diseases and cancers. Their results demonstrated that the AA genotype and A allele of rs231775 were associated with increased risk of multiple types of cancer, such as pancreatic cancer, breast cancer and cervical cancer (10-12,29).
In the present study, we found that rs231775 polymorphisms of CTLA-4 were associated with osteosarcoma risk, and the people with A allele and AA genotype of rs231775 have an increased risk of developing osteosarcoma compared to those G allele carriers. These results were similar to previous reports in some cancers, such as pancreatic cancer, breast cancer and cervical cancer (10-12,29). In addition, there was no significant heterogeneity between studies, suggested the uniform of the subject and method of the included studies. However, all the subjects were Chinese, and no Caucasian ethnicity was included, therefore, our results should be interpreted when extrapolated to other ethnicities.
rs5742909 of CTLA-4 is localized in the promoter region, and the C/T genotype of rs5742909 was associated with higher serum CTLA-4 levels in normal people (30). Recently, Liu et al. (31) showed that the genotype and allele distributions of rs5742909 were differed significantly between RA patients and controls (P<0.05). In the case of cancer, Jiang et al. (29) found that T allele of rs5742909, which is localized in the promoter region, was significantly higher in cervical cancer patients than in controls. In the present study, our results did not support the association between rs5742909 polymorphism and osteosarcoma risk. These results were similar in other cancers, such as lung cancer (32) and melanoma (33).
Although there have been many studies demonstrated the association of TNF-α rs1800629 polymorphism with various cancers (18,20,33), only few studies investigated the role of TNF-α in osteosarcoma. Kawashima et al. (34) showed that TNF-α was able to provoke up-regulation of α2β1 and α5β1 integrins, and cell migration in osteosarcoma cells. Harimaya et al. (5) found that TNF-α was able to induce motility and invasion of human osteosarcoma cells, and this function was possibly through the activation NF-kappa B. Recently, Oliveira et al. (22) showed that osteosarcoma patients with GG genotype of TNF-β rs909253 showed an event-free survival rate of 20% at 100 months. In the present study, our results did not indicate that TNF-α rs1800629 polymorphism conferred a susceptibility to osteosarcoma risk, suggested rs1800629 might to involve in the carcinogenesis of osteosarcoma. However, these results should be interpreted with caution due to the small sample size.
This study clarifies the associations between CTLA-4 or TNF-α polymorphism and the risk of osteosarcoma. We pooled all the available data to explore the genetic association, and the enlarged sample size enhanced statistical power to detect more stable association than any previous individual study. We also assessed the association by testing our results under four different genetic models in order to obtain a robust and accurate estimation. In addition, there were no significant between-study heterogeneity and publication bias in the individual SNP analysis, indicating the reliable of the synthesis results.
However, some limitations of the present study should be noted. First, due to limited studies available, the number of included subjects was relatively small, especially in the analysis of TNF-α. Thus, the statistical power is undermined on this SNP. Second, the ethnicity did not included Caucasian population, and all the subjects were Chinese population in the analysis of CTLA-4, thus, the conclusion may not be applicable to other ethnicities. Third, the present meta-analysis was based on unadjusted evaluation due to lack of the original information, thus, the lack of potential confounder adjustment may raise a concern of bias. Fourth, the limited data also restricted us from evaluating the effect of potential gene-gene interaction on the association. Therefore, in order to provide a more precise evaluation on the basis of adjustment for confounders, a well-designed study is warranted by taking potential confounders or gene-gene interaction into account.
In conclusion, our meta-analysis showed that the rs231775 polymorphism of CTLA-4 was associated to the osteosarcoma risk, while rs1800629 polymorphism of CTLA-4 and rs1800629 polymorphism of TNF-α failed to show the association with the osteosarcoma risk.
Acknowledgments
This work was supported by National Natural Science Foundation (No. 81260315), Foundation of the Education Department of Guangxi Province, China (No. 201010LX375), and the Foundation of the Nature Science Fund, Guangxi Province, China (No. 2012GXNSFBA053121).
Disclosure: The authors declare no conflict interest.
References
- Dorfman HD, Czerniak B. Bone cancers. Cancer 1995;75:203-10. [PubMed]
- Ligers A, Teleshova N, Masterman T, et al. CTLA-4 gene expression is influenced by promoter and exon 1 polymorphisms. Genes Immun 2001;2:145-52. [PubMed]
- Hodi FS, Mihm MC, Soiffer RJ, et al. Biologic activity of cytotoxic T lymphocyte -associated antigen 4 antibody blockade in previously vaccinated metastatic melanoma and ovarian carcinoma patients. Proc Natl Acad Sci USA 2003;100:4712-7. [PubMed]
- Phan GQ, Yang JC, Sherry RM, et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc Natl Acad Sci USA 2003;100:8372-7. [PubMed]
- Harimaya K, Tanaka K, Matsumoto Y, et al. Antioxidants inhibit TNFalpha-induced motility and invasion of human osteosarcoma cells: possible involvement of NFkappaB activation. Clin Exp Metastasis 2000;18:121-9. [PubMed]
- Lesterhuis WJ, Salmons J, Nowak AK, et al. Synergistic effect of CTLA-4 blockade and cancer chemotherapy in the induction of anti-tumor immunity. PLoS One 2013;8:e61895. [PubMed]
- Olson BM, Jankowska-Gan E, Becker JT, et al. Human prostate tumor antigen-specific CD8+ regulatory T cells are inhibited by CTLA-4 or IL-35 blockade. J Immunol 2012;189:5590-601. [PubMed]
- Salvi S, Fontana V, Boccardo S, et al. Evaluation of CTLA-4 expression and relevance as a novel prognostic factor in patients with non-small cell lung cancer. Cancer Immunol Immunother 2012;61:1463-72. [PubMed]
- Contardi E, Palmisano GL, Tazzari PL, et al. CTLA-4 is constitutively expressed on tumor cells and can trigger apoptosis upon ligand interaction. Int J Cancer 2005;117:538-50. [PubMed]
- Lang C, Chen L, Li S. Cytotoxic T-lymphocyte antigen-4 +49G/A polymorphism and susceptibility to pancreatic cancer. DNA Cell Biol 2012;31:683-7. [PubMed]
- Xia W, Shi R, Zheng WL, et al. Lack of association between cytotoxic T-lymphocyte antigen-4 -318C/T polymorphism and cancer risk: a meta-analysis of case-control studies. Technol Cancer Res Treat 2013;12:565-74. [PubMed]
- Zheng J, Yu X, Jiang L, et al. Association between the cytotoxic T-lymphocyte antigen 4 +49G > A polymorphism and cancer risk: a meta-analysis. BMC Cancer 2010;10:522. [PubMed]
- Feng D, Yang X, Li S, et al. Cytotoxic T-Lymphocyte Antigen-4 Genetic Variants and Risk of Ewing's Sarcoma. Genet Test Mol Biomarkers 2013;17:458-63. [PubMed]
- Liu Y, He Z, Feng D, et al. Cytotoxic T-lymphocyte antigen-4 polymorphisms and susceptibility to osteosarcoma. DNA Cell Biol 2011;30:1051-5. [PubMed]
- Wang W, Wang J, Song H, et al. Cytotoxic T-lymphocyte antigen-4 +49G/A polymorphism is associated with increased risk of osteosarcoma. Genet Test Mol Biomarkers 2011;15:503-6. [PubMed]
- Yang S, Wang C, Zhou Y, et al. Cytotoxic T-lymphocyte antigen-4 polymorphisms and susceptibility to Ewing’s sarcoma. Genet Test Mol Biomarkers 2012;16:1236-40. [PubMed]
- Rink L, Kirchner H. Recent progress in the tumor necrosis factor-alpha field. Int Arch Allergy Immunol 1996;111:199-209. [PubMed]
- Hong Y, Ge Z, Jing C, et al. Functional promoter -308G>A in tumor necrosis factor alpha gene is associated with risk and progression of gastric cancer in a Chinese population. PLoS One 2013;8:e50856. [PubMed]
- Berhane N, Sobti RC, Melesse S, et al. Significance of tumor necrosis factor alpha-308 (G/A) gene polymorphism in the development of prostate cancer. Mol Biol Rep 2012;39:11125-30. [PubMed]
- Zhang HL, Zhang YJ. A systemic assessment of the association between tumor necrosis factor alpha 308 G/A polymorphism and risk of cervical cancer. Tumour Biol 2013;34:1659-65. [PubMed]
- Kroeger KM, Carville KS, Abraham LJ. The -308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol 1997;34:391-9. [PubMed]
- Oliveira ID, Petrilli AS, Tavela MH, et al. TNF-alpha, TNF-beta, IL-6, IL-10, PECAM-1 and the MPO inflammatory gene polymorphisms in osteosarcoma. J Pediatr Hematol Oncol 2007;29:293-7. [PubMed]
- Patio-Garcia A, Sotillo-Pieiro E, Modesto C, et al. Analysis of the human tumour necrosis factor-alpha (TNFalpha) gene promoter polymorphisms in children with bone cancer. J Med Genet 2000;37:789-92. [PubMed]
- Xie JT, Liu SQ, Lu BY. Relationship between polymorphism of tumor necrosis factor and osteosarcoma. Zhonghua Shi Yan Wai Ke Za Zhi 2008;25:723-4.
- Gupta R, Prakash S, Parveen F, et al. Association of CTLA-4 and TNF-alpha polymorphism with recurrent miscarriage among North Indian women. Cytokine 2012;60:456-62. [PubMed]
- Han Q, Duan S, Zhang G, et al. Associations between cytotoxic T lymphocyte -associated antigen-4 polymorphisms and serum tumor necrosis factor-alpha and interferon-gamma levels in patients with chronic hepatitis B virus infection. Inflamm Res 2011;60:1071-8. [PubMed]
- Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med 2009;6:e28. [PubMed]
- Ueda H, Howson JM, Esposito L, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003;423:506-11. [PubMed]
- Jiang L, Luo RY, Zhang W, et al. Single nucleotide polymorphisms of CTLA4 gene and their association with human cervical cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2011;28:313-7. [PubMed]
- Torres-Carrillo N, Ontiveros-Mercado H, Torres-Carrillo NM, et al. The -319C/+49G/CT60G Haplotype of CTLA-4 Gene Confers Susceptibility to Rheumatoid Arthritis in Mexican Population. Cell Biochem Biophys 2013;67:1217-28. [PubMed]
- Liu CP, Jiang JA, Wang T, et al. CTLA-4 and CD86 genetic variants and haplotypes in patients with rheumatoid arthritis in southeastern China. Genet Mol Res 2013;12:1373-82. [PubMed]
- Karabon L, Pawlak E, Tomkiewicz A, et al. CTLA-4, CD28, and ICOS gene polymorphism associations with non-small-cell lung cancer. Hum Immunol 2011;72:947-54. [PubMed]
- Breunis WB, Tarazona-Santos E, Chen R, et al. Influence of cytotoxic T lymphocyte-associated antigen 4 (CTLA4) common polymorphisms on outcome in treatment of melanoma patients with CTLA-4 blockade. J Immunother 2008;31:586-90. [PubMed]
- Kawashima A, Kawahara E, Tokuda R, et al. Tumour necrosis factor-alpha provokes upregulation of alpha2beta1 and alpha5beta1 integrins, and cell migration in OST osteosarcoma cells. Cell Biol Int 2001;25:319-29. [PubMed]