Does not hUTP14a promoter form a regulation feedback loop with P53?
Introduction
The U3 proteins (U Three Proteins, UTPs) are found to be part of a large ribonucleoprotein (RNP) complex containing the U3 snoRNA (1) and are part of the active pre-rRNA processing complex. This large RNP complex has been termed the small subunit (SSU) processome (1). We previously identified a member of UTP family, the human UTP14a (hUTP14a) by demonstrating that hUTP14a is nucleolar, associated with U3 small nucleolar RNA, and that knockdown of hUTP14a inhibits 18S rRNA processing (2). In the same study, we also found that hUTP14a caused protein degradation of P53 (2).
P53 is a tumor suppressor protein, which is encoded by the TP53 gene in humans (3-5). The tumor suppressor P53 is mutated in more than 50% of human cancers (6,7). P53 acts as a transcription factor, which can promote cell cycle arrest, apoptosis and senescence via regulating the expression of various downstream genes (8-10). Thus, it is of great importance to keep this protein under tight control. Under normal, non-stressed conditions, P53 is an unstable protein with a short half-life ranging from 5 to 30 min, which is maintained at low level through its predominant regulator the mouse double minute 2 protein (MDM2, or HDM2 for the human ortholog) (11-13). MDM2 is an E3 ligase and promotes P53 degradation through a ubiquitin dependent pathway (14). MDM2 transcription is negatively regulated by P53 and thus MDM2 and P53 forms an autoregulatory negative feedback loop (15). Moreover, several other E3 ligases have been identified, such as P53-induced RING-H2 protein (Pirh2) (16), constitutively photomorphogenic 1 (COP1) (17), and ARF binding protein 1 (ARF-BP1) (18). Among them, P53 activates Pirh2 and COP1 transcription in stressed cells, thus P53 forms a regulation loop with these two P53 E3 ligases, while the transcription of ARF-BP1 is not regulated by P53 (19).
We found that hUTP14a is up-regulated in various types of human cancer tissues (our unpublished data) suggesting that hUTP14 protein might function as an oncogene and play a significant role in tumorigenesis. We thereafter wanted to know if hUTP14a forms a regulation loop with P53 as MDM2 does. To address this issue, we set out to investigate if the transcription of hUTP14a is regulated by P53.
In this study, we defined the promoter of hUTP14a gene and explored the role of P53 on regulating the hUTP14a promoter. These findings will help us to understand the fundamental molecular mechanisms by which hUTP14a functions in the tumorigenesis.
Materials and methods
Bioinformatics
The transcriptional initiation site was identified using the NCBI (http://www.ncbi.nlm.nih.gov/mapview/) and UCSC Genome (http://genome.ucsc.edu/) bioinformatics analysis. Potential binding sites of transcription factors within the promoter region of hUTP14a gene were performed with MatInspector Professional (http://genomatix.de/cgi-bin/matinspector_prof/mat_fam.pl) and Algorismica i Genetica (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/) web softwares.
Cloning of hUTP14a promoter reporter vectors
To obtain the 2,422 bp putative hUTP14a promoter region, a polymerase chain reaction (PCR) on human genomic DNA was performed in a 50 uL reaction mixture. The oligonucleotide sequences of the primers used in the PCR are as following: F-1: 5'-CTAGGAAGCAATGGGACA-3', R-1: 5'-GGCTGGATCGCGGAAAATA-3'. After an initial denaturation step at 94 °C for 5 min, the PCR was carried out for 35 cycles at 94 °C for 15 s, 60 °C for 30 s, and 68 °C for 2 min 30 s, with a final extension at 68 °C for 10 min. The PCR product was purified with glass milk and cloned into pGL3-Basic reporter vector (Promega, Madison, WI) at the Nhe I/Hind III sites. The constructed plasmid pGL3- hUTP14a-luc was verified by DNA sequencing.
Series deletion mutants of hUTP14a promoter were cloned into the pGL3-Basic vector by PCR-cloning as described above to get the following plasmids: pGL3-hUTP14a(-1400/+173)-luc, pGL3-hUTP14a(-745/+173)-luc, pGL3-hUTP14a(-320/+173)-luc, pGL3-hUTP14a(-203/+173)-luc, pGL3-hUTP14a(-100/+173)-luc and pGL3-hUTP14a(-17/+173)-luc. The oligonucleotide sequences used as primers in the PCRs are listed in Table 1. The constructed plasmids were verified by DNA sequencing.
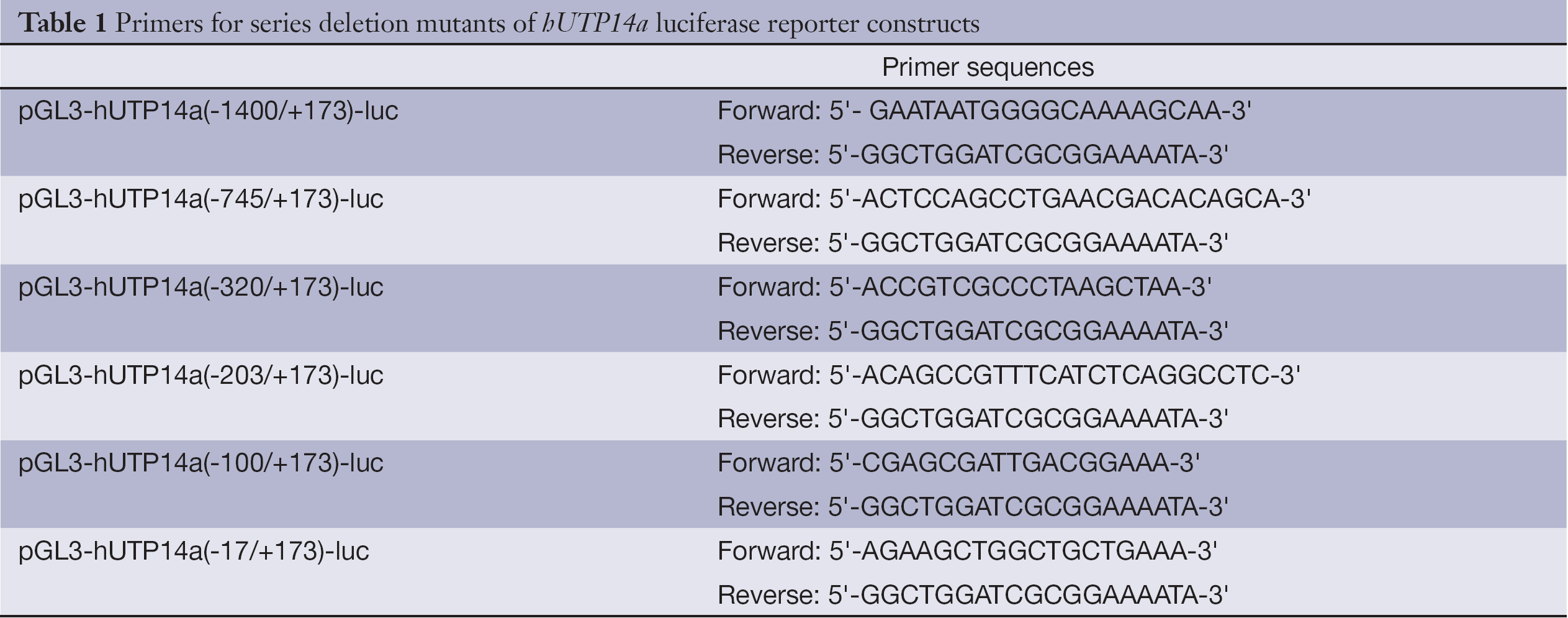
Full table
Cell culture, transient transfection and luciferase assay
U2OS and 293T cells were obtained from the American Type Culture Collection (ATCC) and were grown in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) according to the instructions of the ATCC. Cells were incubated in a humidified atmosphere with 5% CO2 at 37 °C.
Transfections were performed with LipofectamineTM 2000 Reagent (Invitrogen) according to the manufacturer’s instructions. 293T cells were seeded at a density of 1×105 cells per well in a 24-well plate before transfection. Cells were transfected with various hUTP14a promoter constructs or pGL3-basic control plasmid, and a Renilla luciferase control reporter vector (Promega) was used for normalizing transfection efficiency. After 24 h of transfection, the cells were harvested in 100 uL of Passive Lysis Buffer (Promega) and luciferase activity was assayed using the Dual-Luciferase Reporter Assay System (Promega) using Berthold luminometer (Berthold, Wildbad, Germany) according to the manufacturer’s recommendations. Data were presented as relative luciferase activity compared with the pGL3-Basic control, which is normalized to 1.0. Experiments were repeated at least three times in triplicates.
Statistical analysis
All statistical analyses were carried out using SPSS software 17 version (SPSS Inc., Chicago, IL, USA). All data were shown as 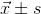 . Data assessing the luciferase activity of hUTP14a promoter were compared using the Student’s t-test, and the identification of hUTP14a minimal promoter was compared with one-way analysis of variance (ANOVA) in different groups. P<0.05 was considered statistically significant.
. Data assessing the luciferase activity of hUTP14a promoter were compared using the Student’s t-test, and the identification of hUTP14a minimal promoter was compared with one-way analysis of variance (ANOVA) in different groups. P<0.05 was considered statistically significant.
Results
Structure analysis and cloning of hUTP14a promoter region
In order to clone the promoter region of hUTP14a, we used the NCBI and UCSC Genome Bioinformatics to analyze the 5' upstream sequence of hUTP14a gene. A database search against the human genomic DNA database was performed using hUTP14a sequence (GenBank accession No. NC_000023) as a query. A genomic DNA fragment that spans position -2248 to +173 including the initiation codon ATG of the hUTP14a gene was chosen as the putative promoter region of hUTP14a gene and this DNA fragment was amplified by PCR (Figure 1A). Figure 1B shows the PCR product which is about 2,500 bp in size. The amplified DNA fragment was cloned into pGL3-Basic reporter vector at the Nhe I/Hind III site (Figure 1C) to get pGL3-hUTP14a-luc plasmid. The constructed plasmid was verified by DNA sequencing (Figure 1D). Figure 1E shows the database search against the human genomic DNA database using the DNA sequence of pGL3-hUTP14a-luc plasmid as a query, which demonstrates that the genomic DNA fragment spanning -2248 to +173 of hUTP14a gene was successfully cloned.
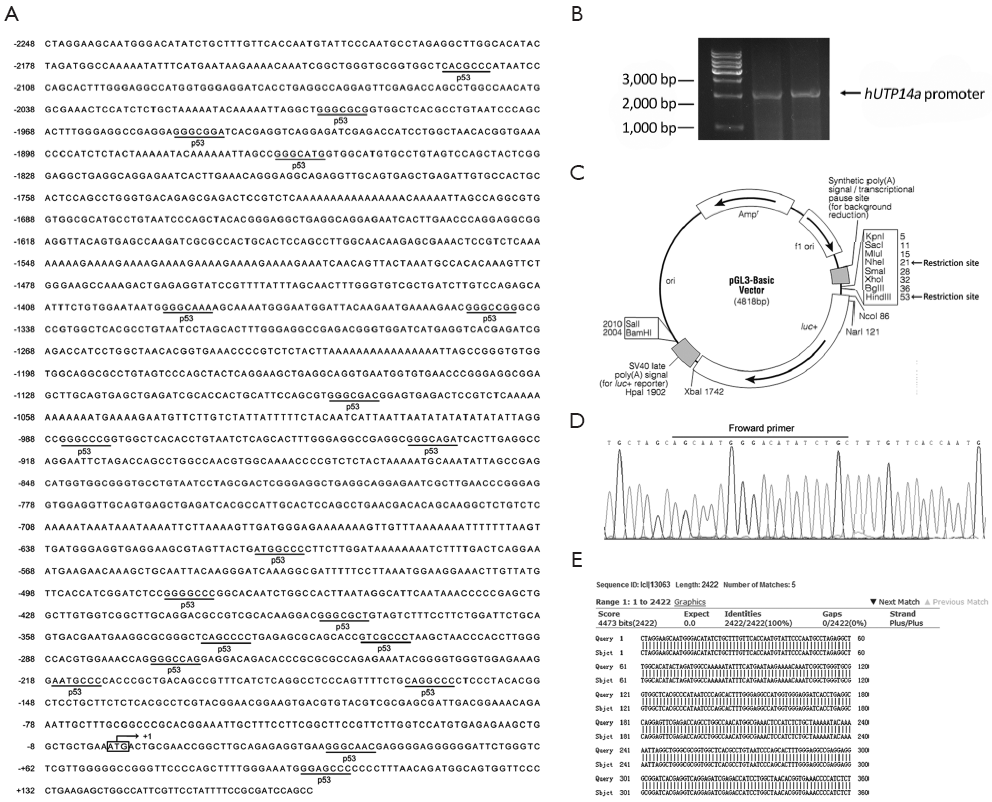
Verification of hUTP14a promoter by luciferase activity
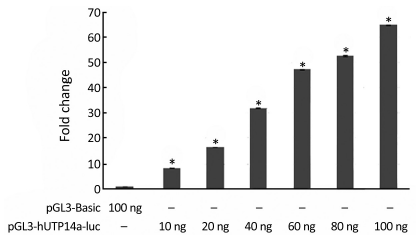
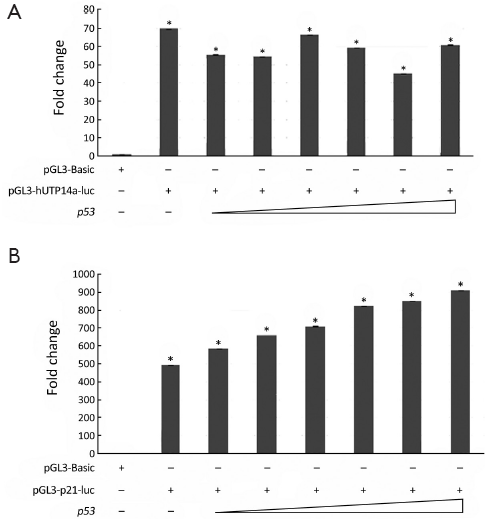
In order to verify if pGL3-hUTP14a-luc reporter construct possesses activity of a promoter, pGL3-hUTP14a-luc plasmid was transiently co-transfected into 293T cells along with Renilla luciferase control reporter vector. The luciferase assay was performed 24 h after transfection. The results showed that the luciferase activity of pGL3-hUTP14a-luc reporter plasmid increased in a dose-dependent manner as compared to the pGL3-Basic empty vector in 293T cells (P<0.05), indicating that an active hUTP14a promoter construct (Figure 2) was obtained.
hUTP14a promoter activity was not regulated by P53
The prediction of transcription factors on hUTP14a promoter revealed a number of putative binding sites for transcription factors. It is a particular interest that P53 was the most frequently found transcription factor on the hUTP14a promoter (Figure 1A), which prompted us to evaluate the transcription regulation of P53 on hUTP14a promoter. To determine the role of P53 in regulating hUTP14a promoter, different amounts of p53 were co-transfected with hUTP14a promoter reporter construct pGL3-hUTP14a-luc into 293T cells. As shown in Figure 3A, the luciferase activity of pGL3-hUTP14a-luc did not show a dose-dependent change when an increasing amount of P53 was ectopically co-expressed. In comparison, the luciferase activity of p21 promoter containing the P53 binding site showed a dose-dependent increase when P53 was co-expressed (Figure 3B). These results indicated that the hUTP14a promoter was not regulated by P53.
Identification of the minimal promoter of hUTP14a
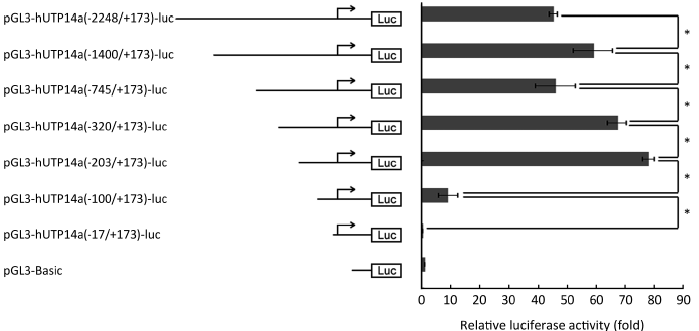
To further confirm that hUTP14a transcription is not regulated by P53 and define the minimal promoter region in hUTP14a promoter, series deletion mutants of hUTP14a promoter reporter plasmid according to the P53 binding sites were constructed as following: pGL3-hUTP14a(-1400/+173)-luc, pGL3-hUTP14a(-745/+173)-luc, pGL3-hUTP14a(-320/+173)-luc, pGL3-hUTP14a(-203/+173)-luc, pGL3-hUTP14a(-100/+173)-luc, and pGL3-hUTP14a(-17/+173)-luc. The diagram of the deletion mutant constructs of hUTP14a promoter reporter is shown in Figure 4 (left). The generated hUTP14a promoter deletion mutant constructs were co-transfected with Renilla luciferase control reporter vector into 293T cells. Luciferase activities were evaluated 24 h after transfection. As shown in Figure 4 (right), the hUTP14a promoter reporter construct and its deletion mutants including pGL3-hUTP14a-luc, pGL3-hUTP14a(-1400/+173)-luc, pGL3-hUTP14a(-745/+173)-luc, pGL3-hUTP14a(-320/+173)-luc and pGL3-hUTP14a(-203/+173)-luc showed fairly high luciferase activities compared to the pGL3-Basic vector. In addition, a significant increase of promoter activity was found between pGL3-hUTP14a-luc and pGL3-hUTP14a(-1400/+173)-luc, pGL3-hUTP14a(-745/+173)-luc and pGL3-hUTP14a(-320/+173)-luc, pGL3-hUTP14a(-320/+173)-luc and pGL3-hUTP14a(-203/+173)-luc, indicating that negative regulatory element(s) may lie between -2248 and -1400, -745 and -203 of hUTP14a gene promoter region. Whereas, a significant decrease of luciferase activity was found between pGL3-hUTP14a(-1400/+173)-luc and pGL3-hUTP14a(-745/+173)-luc, pGL3-hUTP14a(-203/+173)-luc and pGL3-hUTP14a(-100/+173)-luc, suggesting that positive regulatory element(s) may exist between -1400 and -745, -203 and -100 of hUTP14a gene promoter region. These results showed that deleting each of all putative P53 binding sites caused different changes of hUTP14a promoter reporter activity, demonstrating again that hUTP14a transcription is not regulated by P53. It is of notable that the luciferase activity of pGL3-hUTP14a(-100/+173)-luc dropped dramatically compared to that of pGL3-hUTP14a(-203/+173)-luc. This result suggested that the DNA sequence between -203 and -100 to the transcription initiation site of hUTP14a gene is mainly required for the minimal transcription activity and the positive regulatory element for basal transcription activity is mainly located in the region between -203 and -100.
Discussion
Our previous study showed that hUTP14a expression caused P53 protein degradation, knockdown of hUTP14a inhibits cell growth and induces apoptosis (2). In addition, hUTP14a is up-regulated in various types of human cancer tissues (our unpublished data). However, little is known about the transcriptional mechanisms controlling hUTP14a gene expression. To find out the mechanisms of hUTP14a gene transcriptional regulation, we first constructed the hUTP14a promoter.
The 2,422 bp fragment, which was originally obtained from human genomic DNA by PCR, was found to contain the promoter region of the hUTP14a gene. The hUTP14a promoter luciferase reporter construct showed a dose-dependent luciferase activity, indicating that the hUTP14a promoter we obtained possesses an active promoter activity.
Our previous study found that hUTP14a binds P53 and promotes P53 degradation (2). Given that the transcription of MDM2 (14), Pirh2 (16) and COP1 (17), which are P53-binding proteins and promote P53 degradation, is regulated by P53, therefore these proteins form an autoregulatory feedback loop with P53. In addition, bioinformatics analysis of transcription factors’ binding sites on hUTP14a promoter revealed several potential P53 binding sites. We thus wanted to know if P53 regulates hUTP14a promoter. hUTP14a promoter was transfected into 293T cells with p53 and pGL3-Basic control plasmid, and luciferase activity was determined. To our surprise, the result showed that ectopic expression of P53 failed to regulate the hUTP14a promoter activity. In comparison, the luciferase activity of p21 promoter containing the P53 binding site was stimulated by P53 in a dose-dependent way. Collectively, these data indicate that the hUTP14a promoter is not regulated by P53.
In order to define the minimal regulatory region in the hUTP14a promoter and to confirm that hUTP14a transcription is not regulated by P53, we generated a series of deletion mutants of the hUTP14a luciferase reporter construct according to the position of putative P53 binding sites. The results of luciferase activity indicated that negative regulatory element(s) may lie in the two regions, one is the region between -2248 and -1400, the other is between -745 and -203 of hUTP14a gene promoter, while positive regulatory element(s) may exist in the regions including the DNA fragment from -1400 to -745 and that from -203 to -100 of hUTP14a gene promoter region. Deletion of each putative P53 binding sites displayed different luciferase activity changes, which demonstrated again that hUTP14a transcription is not regulated by P53. It is of importance that a 103 bp region (-203 to -100) was found to be the uppermost element for the transcriptional activity of the hUTP14a promoter. Further study is to identify specific transcriptional regulation factors of hUTP14a in the promoter region between -203 to -100.
Several specific E3 ubiquitin ligases such as MDM2 (14), Pirh2 (16,20), COP1(17,21) and ARF-BP1 (18) play as E3 ligases for P53 and mediate P53 degradation through an ubiquitin-dependent pathway. Among them, transcriptions of MDM2, Pirh2 and COP1 are regulated by P53 and thus these E3 ligases form a feedback regulation loop with P53 when cells are under stressed conditions (19). However, the transcription of ARF-BP1 is not regulated by P53 under either unstressed or stressed conditions so far (19). We previously found that hUTP14a failed to promote P53 polyubiquitination though it promotes a proteasomal degradation of P53 (2). In this study, we demonstrate that transcription of hUTP14a is not regulated by P53 under unstressed condition. Whether P53 regulates hUTP14a transcription under stressed conditions needs further study.
The other possibility that P53 has no effect on hUTP14a promoter can also be attributed to the cell lineage. There is endogenous expression in 293T cells, which may influence the interaction between the exogenous P53 and hUTP14a promoter. Human Saos2 osteosarcoma cells, which harbor a homozygous deletion of the P53 locus and do not produce P53 protein, will be used in the further study to investigate whether P53 functions on the hUTP14a promoter. In addition, wild-type p53 or mutant p53 along with hUTP14a promoter construct will be co-transfected into the human Saos2 cells to see if P53 regulates hUTP14a promoter.
In conclusion, we cloned the hUTP14a promoter and defined its minimal promoter. The hUTP14a promoter is not regulated by P53 under unstressed conditions. The essential transcriptional factors of hUTP14a gene may exist in the promoter region between -203 and -100. Further understanding of the transcriptional regulation of the hUTP14a gene will provide important information about functions of hUTP14a in tumorigenesis.
Acknowledgements
This work was supported by grants from Beijing Municipal Natural Science Foundation (Grant No. 7122032), the National Natural Science Foundation of China (Grant No. 81071672), the National Basic Research Program of China (973 program) (Grant No. 2010CB529303) and National High Technology Research and Development Program of China (863 Program, Grant No. 2008AA02Z131).
Disclosure: The authors declare no conflict of interest.
References
- Dragon F, Gallagher JE, Compagnone-Post PA, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002;417:967-70. [PubMed]
- Hu L, Wang J, Liu Y, et al. A small ribosomal subunit (SSU) processome component, the human U3 protein 14A (hUTP14A) binds p53 and promotes p53 degradation. J Biol Chem 2011;286:3119-28. [PubMed]
- Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2010;2:a000893. [PubMed]
- Matlashewski G, Lamb P, Pim D, et al. Isolation and characterization of a human p53 cDNA clone: expression of the human p53 gene. EMBO J 1984;3:3257-62. [PubMed]
- McBride OW, Merry D, Givol D. The gene for human p53 cellular tumor antigen is located on chromosome 17 short arm (17p13). Proc Natl Acad Sci U S A 1986;83:130-4. [PubMed]
- Garritano S, Inga A, Gemignani F, et al. More targets, more pathways and more clues for mutant p53. Oncogenesis 2013;2:e54. [PubMed]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15:2-8. [PubMed]
- Hasty P, Christy BA. p53 as an intervention target for cancer and aging. Pathobiol Aging Age Relat Dis 2013;3.
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000;408:307-10. [PubMed]
- Lee CL, Blum JM, Kirsch DG. Role of p53 in regulating tissue response to radiation by mechanisms independent of apoptosis. Transl Cancer Res 2013;2:412-21. [PubMed]
- Nag S, Qin J, Srivenugopal KS, et al. The MDM2-p53 pathway revisited. J Biomed Res 2013;27:254-71. [PubMed]
- Zhou X, Liao JM, Liao WJ, et al. Scission of the p53-MDM2 loop by ribosomal proteins. Genes Cancer 2012;3:298-310. [PubMed]
- Hamzehloie T, Mojarrad M, Hasanzadeh Nazarabadi M, et al. The role of tumor protein 53 mutations in common human cancers and targeting the murine double minute 2-p53 interaction for cancer therapy. Iran J Med Sci 2012;37:3-8. [PubMed]
- Ponnuswamy A, Hupp T, Fåhraeus R. Concepts in MDM2 signaling: allosteric regulation and feedback loops. Genes Cancer 2012;3:291-7. [PubMed]
- Haupt Y, Maya R, Kazaz A, et al. MDM2 promotes the rapid degradation of p53. Nature 1997;387:296-9. [PubMed]
- Jung YS, Qian Y, Chen X. Pirh2 RING-finger E3 ubiquitin ligase: its role in tumorigenesis and cancer therapy. FEBS Lett 2012;586:1397-402. [PubMed]
- Lee YH, Andersen JB, Song HT, et al. Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer Res 2010;70:8264-9. [PubMed]
- Chen D, Kon N, Li M, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 2005;121:1071-83. [PubMed]
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell 2006;21:307-15. [PubMed]
- Leng RP, Lin Y, Ma W, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003;112:779-91. [PubMed]
- Dornan D, Wertz I, Shimizu H, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004;429:86-92. [PubMed]
