Association between hsa-miR-34b/c rs4938723 T > C promoter polymorphism and cancer risk: a meta-analysis based on 6,036 cases and 6,204 controls
Introduction
MicroRNAs (miRNAs) are non-coding RNAs of approximately 21 to 25 nucleotides in length. Mature miRNAs target the 3' untranslated region of mRNA, thereby degrading mRNA or suppressing translation (1,2). A single miRNA can regulate multiple genes implicated in multiple pathways (3), and a signaling pathway can be affected by a series of miRNAs (4,5). Therefore, miRNAs have key functions in gene network regulation as well as in physiological and pathological processes, including tumorigenesis, proliferation, apoptosis, and metabolism (1,3,6-8).
MiR-34b and miR-34c share a common primary transcript (pri-miR-34b/c). These miRNAs are frequently downregulated and function as tumor suppressors in several types of cancer. For instance, the miR-34 family members are direct transcriptional targets of p53 and constitute a part of the p53 tumor suppressor network. These family members also regulate cell cycle arrest, apoptosis, and senescence (9,10). Microarray analyses have revealed that the miR-34 family can down-regulate numerous putative target genes, including CDK4/6, Cyclin E2, MET, and Bcl-2, which have important functions in tumor development (9-11). MiRNA-34b can also inhibit prostate cancer via demethylation, active chromatin modifications, and AKT pathways (12).
Pri-miR-34b/c contains a corresponding promoter. Single nucleotide polymorphisms (SNPs) in the promoter region may be important in the alteration of expression and/or function of mature miR-34b/c and its targeted genes, thereby affecting tumor-related pathways. A potentially functional polymorphism (rs4938723 T/C) has been discovered in the promoter region of pri-miR-34b/c by in silico analysis. The T to C shift of the rs4938723 polymorphism possibly influences GATA-X binding sites. If the polymorphic location is C, then it can bind to the GATA-X; otherwise, it cannot bind to the GATA-X (13). Studies have identified that the genetic variants in the pri-miRNA sequence of miR-34b/c (rs4938723 T/C) can be used as possible biomarkers of cancer development. However, inconsistent and inconclusive results have been reported probably because of the slight effect of polymorphism on cancer risk or the relatively small sample size in published studies. Therefore, we conducted this meta-analysis to summarize the effect of the miR-34b/c polymorphism on cancer risk.
Materials and methods
Publication search
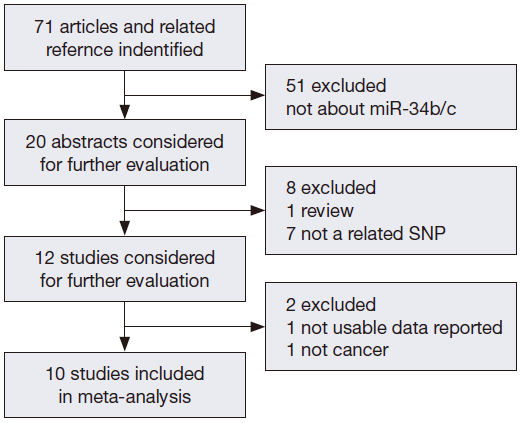
We searched the PubMed, Web of Science, and Chinese National Knowledge Infrastructure (CNKI) databases (last updated on May 1th, 2014) for published studies on the relationship of miR-34b/c polymorphism with cancer risk. The following keywords were used either separately or together: “miR-34b/c or rs4938723”; “polymorphism or mutation”; and “cancer or tumor”. The references of related studies were reviewed to search other potentially related articles manually. All of the selected studies in our meta-analysis should also match the following inclusion criteria: (I) human case-control study; (II) evaluation of miR-34b/c rs4938723 polymorphism and cancer risk; and (III) availability of genotype frequencies in the study. We also restricted our search to studies published in English and Chinese. The procedures of database search are shown in Figure 1.
Data extraction
Two investigators independently obtained the following information from each study: (I) name of the first author; (II) year of publication; (III) country of origin; (IV) ethnicity; (V) cancer type; (VI) total number of control subjects and cases; (VII) genotype frequencies for cases and control subjects; and (VIII) Hardy-Weinberg equilibrium (HWE) of control subjects. Differences in the findings of the investigators were resolved by discussion.
Statistical analysis
HWE was calculated for the control subjects of each study by using a Chi-square goodness-of-fit test. Studies with inconsistent HWE (P<0.05) were removed. The STATA software (version 12.0; Stat Corporation, College Station, Texas, USA) was used to conduct the statistical analysis; all of the tests were two-sided. The odds ratio (OR) and 95% confidence interval (CI) were used to evaluate the strength of the association between the miR-34b/c polymorphism and cancer risk. We determined the relationship between SNP and cancer susceptibility by using the following genetic models: homozygote model (CC/TT); heterozygote model (CT/TT); the dominant genetic model (CCCT/TT); and recessive genetic model (CC/CTTT). Subgroup analyses were also conducted according to cancer type and ethnic group. The Z test was used to determine the significance of the pooled OR, in which the results were considered statistically significant at P<0.05. The heterogeneity among studies was determined by using a Chi-square-based Q test. The fixed-effect model (Mantel-Haenszel model) was used when homogeneity was considered significant (P>0.05); otherwise, the random-effect model (DerSimonian and Laird method) was used. In addition, publication bias was assessed using Begg’s funnel plot and Egger’s test (P<0.05 was considered a significant publication bias). These procedures were conducted twice in our meta-analysis.
Results
Characteristics of the studies
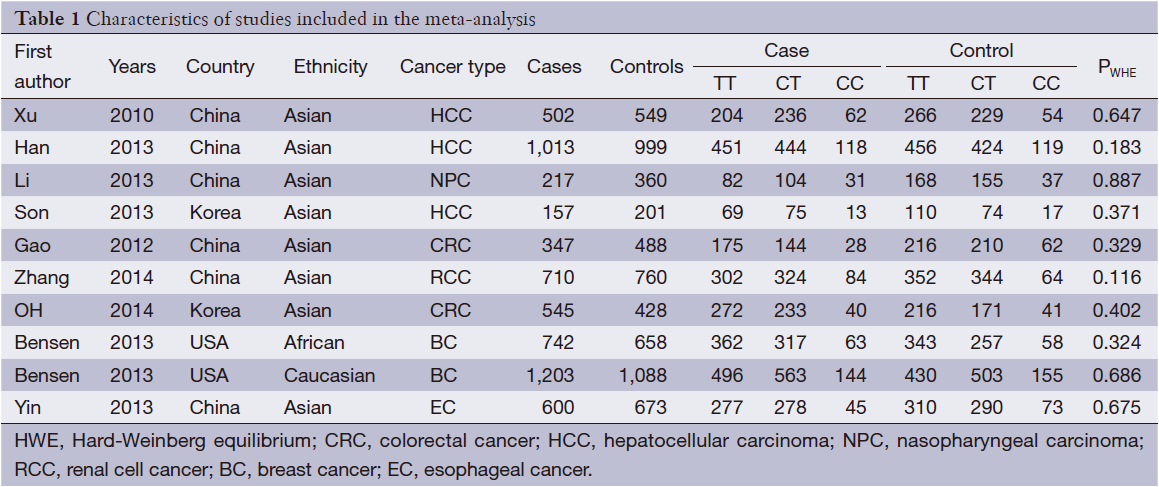
After an extensive search was conducted, a total of ten studies (with 6,036 cancer patients and 6,204 control subjects) satisfied our criteria. The details of each study are shown in Table 1. Among these studies, three focused on hepatocellular carcinoma (HCC), two focused on colorectal cancer (CRC), two focused on breast cancer (BC) and the other three focused on other types of cancers (13-21). The genotypic distributions in the control populations were consistent with HWE in the selected studies.
Full table
Quantitative synthesis and heterogeneity analysis
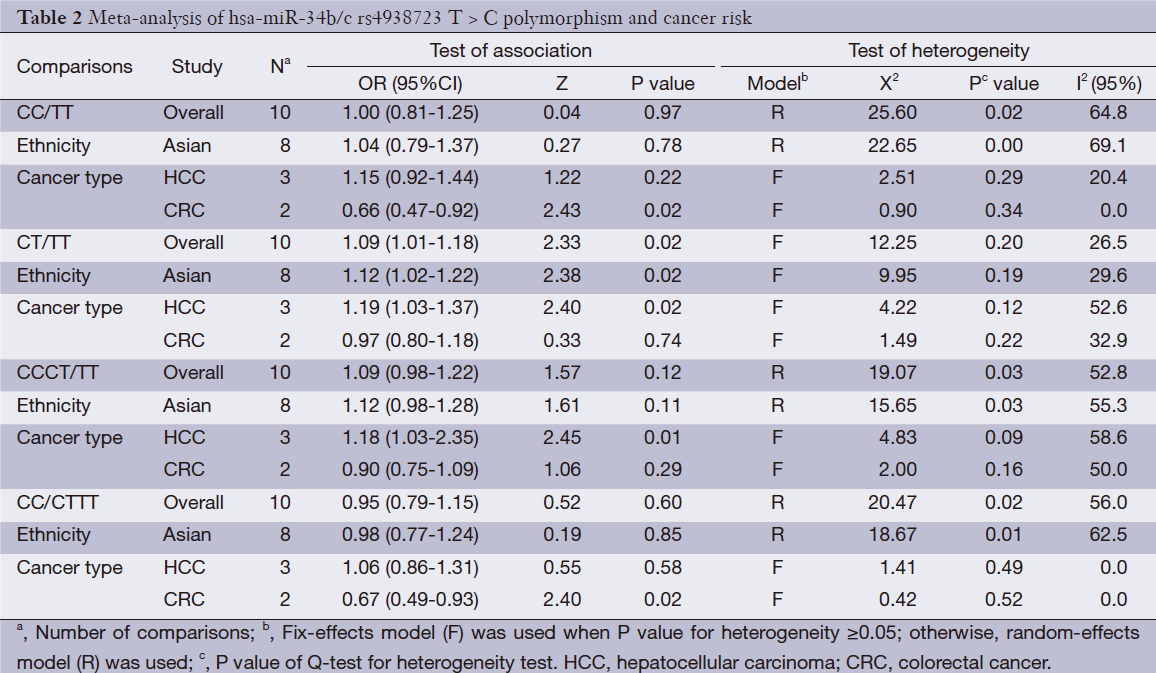
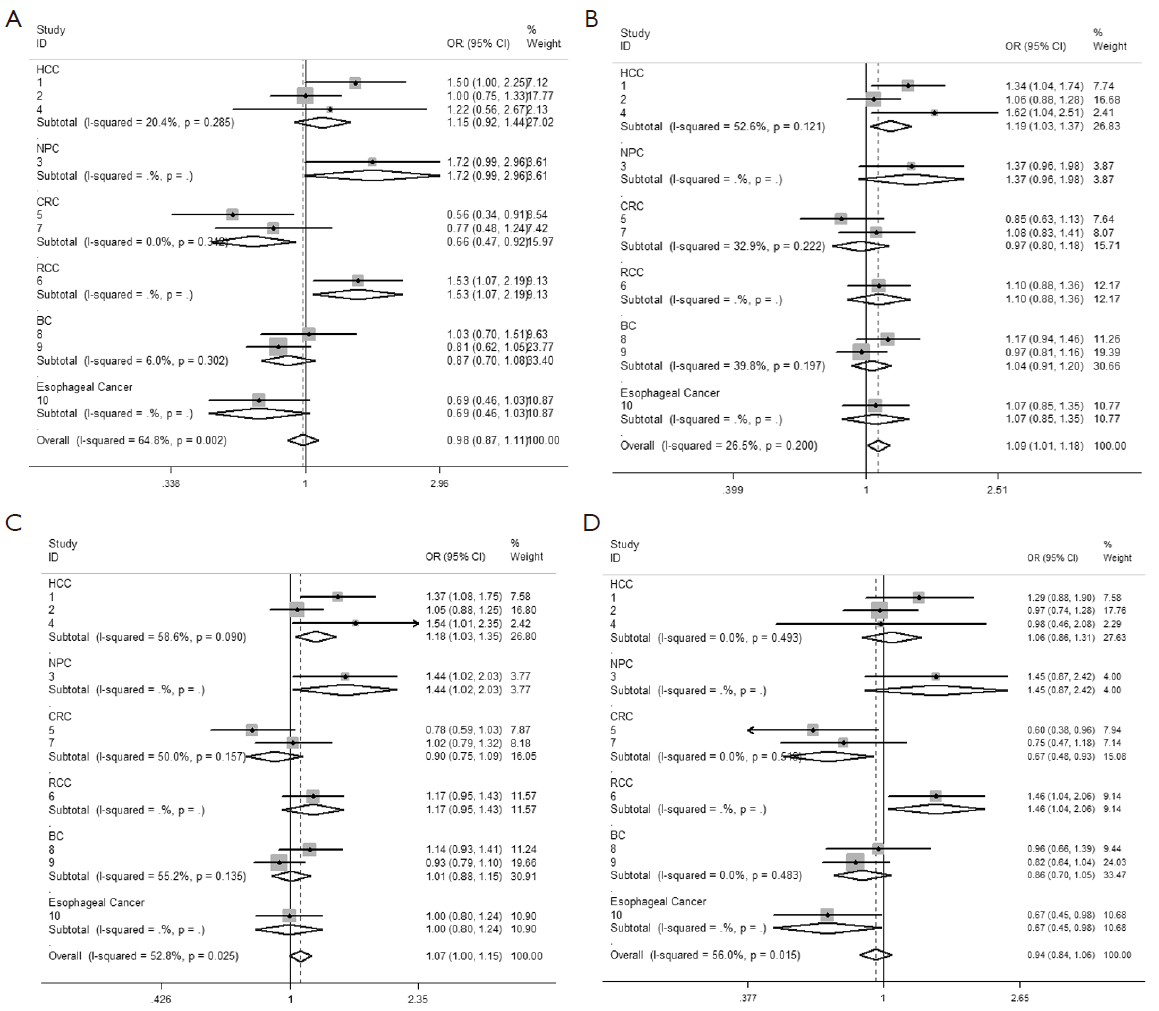
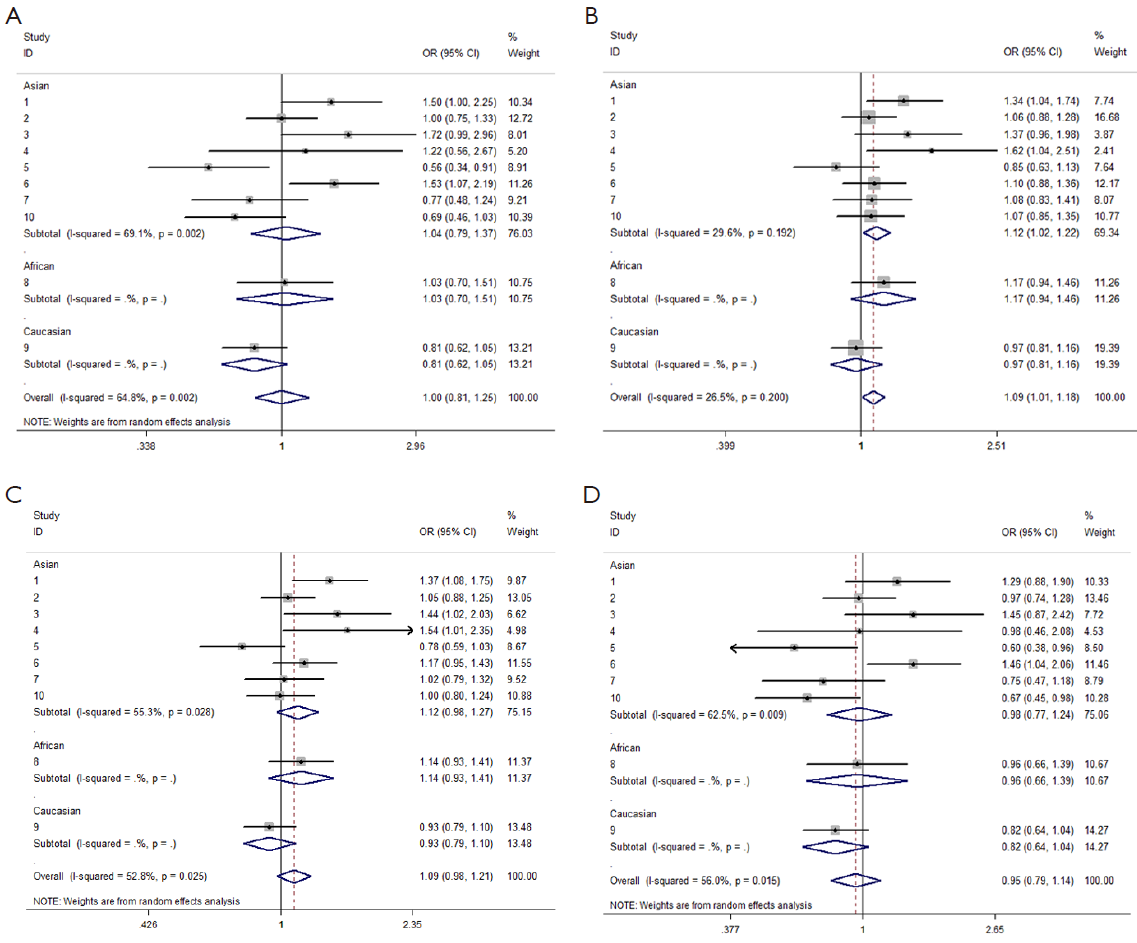
We observed that hsa-miR-34b/c rs4938723 T > C polymorphism was weakly associated with overall cancer susceptibility in the variant CT genotype (Table 2 and Figure 2) after we pooled the selected studies in the meta-analysis. The Q-test of heterogeneity was always significant, and we conducted analyses using random effect models for the overall population if Pheterogeneity<0.05. Then, a stratified analysis was performed according to cancer type. The results showed that the variant CT (OR =1.19, 95% CI: 1.03-1.37) and CC/CT (OR =1.18, 95% CI: 1.03-2.35) genotypes were associated with an increased risk of HCC compared with wild-type TT genotype. However, a decreased risk of CRC was found in the genetic model of CC/TT (OR =0.66, 95% CI: 0.47-0.92) and CC/CTTT (OR =0.67, 95% CI: 0.49-0.93) (Figure 2). We also performed the analysis stratified by ethnicity. The results indicated that the variant CT (OR =1.12, 95% CI: 1.02-1.22) genotype were associated with an increased risk of cancer compared with wild-type TT in Asian population (Figure 3). Base on the difference of OR value in subgroups, we thought that different cancer types were the main origin of heterogeneity.
Full table
Publication bias
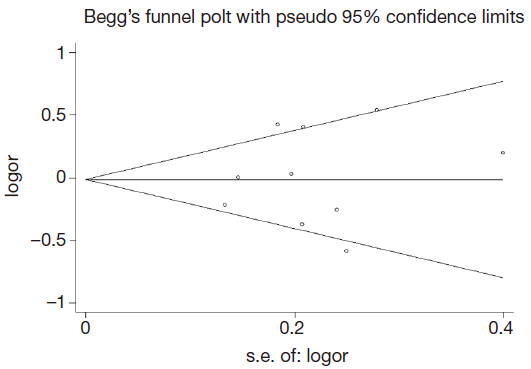
Publication bias was assessed by Begg’s funnel plot and Egger’s test (Figure 4). We found that the graphical funnel plots were symmetrical for the comparison of the genetic models. Egger’s test results did not indicate any evidence of publication bias in our meta-analysis (P>0.05).
Sensitivity analysis
Sensitivity analyses were performed after sequential removal of each eligible study. The results suggested that the significance of the pooled ORs in overall analysis was influenced in heterozygote model by three studies conducted by Yan Xu et al., Myung Su Son et al., and Jeannette T Bensen et al. (14,19,20). The pooled OR and CI values were effectively changed by exclusion of the three studies: OR =1.08 (95% CI: 0.99-1.18), 1.08 (95% CI: 1.00-1.17), and 1.08 (95% CI: 1.00-1.18) after removal, respectively. The significance of the ORs in the analysis stratified by ethnicity was influenced in heterozygote model by Yan Xu et al.’s study (14). The pooled OR and CI values were weakly changed by exclusion of this study: OR =1.09 (95% CI: 0.99-1.20) after removal. Therefore, the positive result about heterozygote model and cancer risk in Figure 2 and Figure 3 and Table 2 was needed to be treated with caution. Based on the above data, lack of association was found between hsa-miR-34b/c rs4938723 polymorphism and overall cancer susceptibility. Further, the sensitivity analysis suggested that the significance of the pooled ORs was not influenced by any single study in other genetic models (data not shown).
Discussion
Evidence indicates that a complex network of “miRNA-mRNA” is involved in human cancer (22). However, the mechanisms regulating miRNA expression and function have not been elucidated. SNPs in miRNA genes can possibly affect gene expression and function by modulating the transcription of the primary transcript (pri-miRNA) as well as the processing and maturation of pre-miRNA. Studies have shown that pri-miRNAs or pre-miRNA SNPs are associated with cancer risk. For instance, rs7372209 in pri-miR-26a-1 is associated with a 64% decreased risk of bladder cancer in females and a twofold increased risk of premalignant oral lesions (23,24). The rs531564 SNP in pri-miR-124-1 is also associated with increased risks of bladder cancer and esophageal cancer (EC) in males (25). Furthermore, the pri-miRNA SNP rs213210 in miR-219 is associated with an increased the risk of EC. pre-miR-196a2 rs11614913 polymorphism is also associated with a significant increased risk of BC (26).
In our meta-analysis, lack of association was found between the hsa-miR-34b/c rs4938723 polymorphism and cancer susceptibility based on ten case-control studies including 6,036 cancer patients and 6,204 control subjects. Subgroup analysis revealed that the variant CT and CC/CT genotypes were associated with an increased risk of HCC compared with wild-type TT genotype. However, a decreased risk of CRC was found in the homozygote genetic model and the recessive genetic model. Recently, CpG methylation of miR-34b/c has been detected in varieties of human cancers. There may be a possibility that the methylation status of miR-34b/c is different between HCC and CRC, which results in the different effect of the miR-34b/c rs4938723 polymorphism on the risk of HCC and CRC. Further large-scale studies, however, should be done to improve the generalization of the results (13,27). Our meta-analysis showed several limitations. First, cancer is caused by many factors, such as genetic disorders and environmental damage, while we only focused on the function of SNP in this study. Second, cancer patients from the studies of Han et al. and Son et al. (15,19) carried hepatitis B/C virus, which may interact with pre-miR-34b/c and influence the results. Third, the lack of original data, including age, gender, and control source, limited our analysis. Fourth, only studies published in English and Chinese were included in our meta-analysis. This language criterion may restrict our sample size.
Despite these limitations, our meta-analysis reveals that there is no association between hsa-miR-34b/c rs4938723 polymorphism and overall cancer risk based on current studies. Further, this polymorphism plays a different role in the risk of HCC and CRC. To confirm our results, we recommend that future well-designed studies and large sample size should consider diverse ethnic populations and cancer types.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81370849, 81300472, 81070592 and 81202034), Natural Science Foundation of Jiangsu Province (BL2013032 and BK2012336) and Nanjing City (201201053) and Southeast University (3290002402), Science Foundation of Ministry of Education of China (20120092120071), Fundamental Research Funds for the Central Universities and Graduate Innovative Fundation of Jiangsu Province (CXZZ13_0133).
Disclosure: The authors declare no conflict of interest.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [PubMed]
- Gong J, Zhang JP, Li B, et al. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 2013;32:3071-9. [PubMed]
- Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet 2005;37:495-500. [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [PubMed]
- Mocellin S, Pasquali S, Pilati P. Oncomirs: from tumor biology to molecularly targeted anticancer strategies. Mini Rev Med Chem 2009;9:70-80. [PubMed]
- Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008;451:1125-9. [PubMed]
- Aumiller V, Förstemann K. Roles of microRNAs beyond development--metabolism and neural plasticity. Biochim Biophys Acta 2008;1779:692-6.
- He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007;447:1130-4. [PubMed]
- Hermeking H. p53 enters the microRNA world. Cancer Cell 2007;12:414-8. [PubMed]
- Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 2007;17:1298-307. [PubMed]
- Majid S, Dar AA, Saini S, et al. miRNA-34b inhibits prostate cancer through demethylation, active chromatin modifications, and AKT pathways. Clin Cancer Res 2013;19:73-84. [PubMed]
- Gao LB, Li LJ, Pan XM, et al. A genetic variant in the promoter region of miR-34b/c is associated with a reduced risk of colorectal cancer. Biol Chem 2013;394:415-20. [PubMed]
- Xu Y, Liu L, Liu J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer 2011;128:412-7. [PubMed]
- Han Y, Pu R, Han X, et al. Associations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma risk. PLoS One 2013;8:e58564. [PubMed]
- Li L, Wu J, Sima X, et al. Interactions of miR-34b/c and TP-53 polymorphisms on the risk of nasopharyngeal carcinoma. Tumour Biol 2013;34:1919-23. [PubMed]
- Zhang S, Qian J, Cao Q, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with renal cell cancer risk in a Chinese population. Mutagenesis 2014;29:149-54. [PubMed]
- Oh J, Kim JW, Lee BE, et al. Polymorphisms of the pri-miR-34b/c promoter and TP53 codon 72 are associated with risk of colorectal cancer. Oncol Rep 2014;31:995-1002. [PubMed]
- Son MS, Jang MJ, Jeon YJ, et al. Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene 2013;524:156-60. [PubMed]
- Bensen JT, Tse CK, Nyante SJ, et al. Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: the Carolina Breast Cancer Study. Cancer Causes Control 2013;24:1099-109. [PubMed]
- Yin J, Wang X, Zheng L, et al. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLoS One 2013;8:e80570. [PubMed]
- Tao T, Wang Y, Luo H, et al. Involvement of FOS-mediated miR-181b/miR-21 signalling in the progression of malignant gliomas. Eur J Cancer 2013;49:3055-63. [PubMed]
- Yang H, Dinney CP, Ye Y, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res 2008;68:2530-7. [PubMed]
- Carlquist JF, Horne BD, Mower C, et al. An evaluation of nine genetic variants related to metabolism and mechanism of action of warfarin as applied to stable dose prediction. J Thromb Thrombolysis 2010;30:358-64. [PubMed]
- Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460-9. [PubMed]
- Gao LB, Bai P, Pan XM, et al. The association between two polymorphisms in pre-miRNAs and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 2011;125:571-4. [PubMed]
- Vogt M, Munding J, Grüner M, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch 2011;458:313-22. [PubMed]