Deletion and down-regulation of SMAD4 gene in colorectal cancers in a Chinese population
Introduction
Colorectal cancer (CRC) has traditionally been one of the most common malignant disorders in western populations. However, during the past few decades, there have been remarkable changes in the incidence of CRC in Asian countries (1). In China, CRC is one of the three cancers with most rapidly increasing incidence (together with lung cancer and female breast cancer). Lifetime risk in the Chinese population is about 4.2% (1,2). CRC is a genetic disease in which the clonal accumulation of genetic alterations confers a cell from adenomatous polyps with the malignant characteristics of uncontrolled growth, local invasiveness, and metastatic potential (3). CRC has a complex pathogenesis involving multiple sequential steps with an accumulation of genetic alterations including mutations, gene amplification, and epigenetic changes (4-6).
Transforming growth factor-beta (TGF-beta) family members exert their function via specific type I and type II serine/threonine kinase receptors and intracellular Smad transcription factors, including the common mediator SMAD4. The dual effects of TGF-beta signaling on tumor initiation and progression are cell-specific and yet to be determined under distinct contexts (7,8). A number of genetically manipulated mouse models with alterations in the TGF-beta pathway genes, particularly the pivotal SMAD4, revealed that these genes play crucial functions in maintaining tissue homeostasis and suppressing tumorigenesis. SMAD4 was initially isolated as a tumor suppressor gene on chromosome 18q21.1 in pancreatic ductal adenocarcinomas (9). It is a central downstream regulator in the transforming growth factor-β (TGF-β) signaling pathway, which forms hetero-oligomeric complexes with activated SMAD 2/3 and regulates gene expression (8,10,11). SMAD4 inactivation at the gene or protein level has been shown to be essential for the progression of several types of tumor including CRC (12-16). Importantly, studies revealed that SMAD4 homozygous deletion and loss of heterozygosity were two of the major mechanisms for the inactivation (17,18) and statistic data showed that SMAD4 has been mutated or deleted in about 15% of colorectal tumors (19). In China, however, limited data about the abnormalities of the SMAD4 gene in a large number of CRC patients has been reported.
Most of the array comparative genomic hybridization (aCGH) experiments focused on the genome-wide screening of copy number variations (CNVs) and the data obtained are generally informative but not definitive. Thus, a study comprehensively examining CNVs in relation to SMAD4 expression or prognosis should be performed using a large number of tumors. In the present study, we collected 147 sporadic CRC samples with matched adjacent normal tissues (ANTs) from Chinese population for CNV analysis. Copy number deletions of SMAD4 were present (34.6%) in CRC samples. There was a positive correlation between copy number loss of SMAD4 and mRNA down-regulation. These findings suggested the potential role of CNVs of SMAD4 in sporadic CRCs.
Materials and methods
Patients and controls tissue collection
CRC samples were obtained from 147 surgical patients of the Department of Gastroenterology, Zhongshan Hospital, Wuhan University between April 2010 to Dec 2013. The self-paired adjacent normal mucosa samples, located at least 2 cm far from the macroscopically unaffected margins of the tumor (polyp or carcinoma), were defined as normal controls. All collected samples were stored in liquid nitrogen. The tissue samples were obtained with informed consent and used with approval from the ethical committee of Wuhan University. The patients’ pathology slides were analyzed according to tumor size, histologic grading, depth of invasion, and presence of nodal metastasis. CRC samples were staged according to the Dukes classification system: Dukes A (T1-T2, N0, and M0; n=49), Dukes B (T3-T4, N0, and M0; n=38), Dukes C (any T, N1-2, M0; n=52), and Dukes D (any T and any N and M1; n=8). Because the samples of Dukes D were limited, we combined these with Dukes C as Dukes C/D.
DNA extraction and quantification of copy numbers
Genomic DNA of patients and controls were isolated from the tissues using the Genomic DNA Extraction Kit (Innogent, Shenzhen, China) according to the manufacturer’s instructions. Matched samples of CRC and normal colonic mucosa (n=147) were subjected to real-time PCR analysis. Meanwhile, genomic DNA from normal peripheral blood (n=152) was also prepared for analyzing the CNVs of SMAD4 with self-paired normal control tissues. Quantitative PCR was performed through BioRad Chromo4 real-time PCR system. Average copy numbers of RNAse P in normal candidates (copy numbers =2) were used as control (20). For relative quantification, the reactions were performed in a total volume of 20 µL that included 12.5 µL of 2xIQTMSYBR Green® Supermix (Bio-Rad, USA), 1 µL of DNA (10 ng/µL), and 1 µL of each primer (10 pmol/µL). The primers for RNAse P are: forward: 5'-AGA CTA GGG TCA GAA GCA A-3' and reverse: 5'-CAT TTC ACT GAA TCC GTT C-3'. The primers for SMAD4 are: forward: 5'- AAG GCC TAG CAC CAC CTT AG-3' and reverse: 5'-AGC CTT AAA CTC TGA CCT GT-3'. The following cycling conditions were employed: initial denaturation 95 °C 3 min, followed by 40 cycles of 95 °C 30 seconds, 58 °C 30 seconds, 72 °C 30 seconds, plate read. A final extension of 72 °C 10 minutes and melting curve of 55 to 95 °C, 1 °C/second transition, were incorporated. Raw data were exported to Microsoft Excel for analysis. The copy numbers of SMAD4 was calculated by using the comparative C(T) method. Cut-off values of 0.33, 0.67, 1.33, and 1.67 were used to define the copy numbers as 0, 1, 2, and 3, respectively. Statistical analysis was performed using chi-square test or Fisher exact test. The significant level was set at 0.05. A representative diagram for the DNA CNV analysis of the target gene in three CRC/ANT pair was shown in Figure 1.

RNA extraction and quantitative RT-PCR
Total RNA was isolated from tissues by using the AxyPrepTM blood total RNA MiniPrep Kit (Axygen) according to the manufacturer’s instructions. First strand cDNA was synthesized with the RevertAidTM first stand cDNA synthesis kit (Fermentas). Quantitative PCR was performed through the BioRad Chromo4 real-time PCR system. At the end point of PCR cycles, melt curves were made to check product purity. Since the efficiencies of amplification of targeted genes were very near to 100%, we used 2-∆ct method to calculate real-time PCR results. The mRNA level of SMAD4 was expressed as a ratio relative to the GAPDH mRNA in each sample. Exploratory data analysis using box plot was applied to visually identify the expression level of target mRNA.
Statistical analysis
Statistical analysis was performed with the SPSS Software (version 12). Data were analyzed by the chi-square test or Fisher exact test. P values less than 0.05 were considered statistically significant. Results of the SMAD4 mRNA expression for normal and tumor tissue samples were compared using two-way repeated measurement ANOVA. One-way, repeated measures analysis of variance (ANOVA-RM) was performed at a significance level of P=0.05 to determine differences from controls within each group. Two-way analysis of variance (ANOVA-2) was performed after baseline subtraction, at a significance level of P=0.05 to determine differences between the groups with deleted and unaltered SMAD4 copy number.
Results
Gene copy number loss of SMAD4 in CRC samples
As shown in Table 1, no statistical differences of copy number distribution between ANTs and healthy normal controls (HNCs) were observed, thus the ANT could be used as controls for the CRC tissues in the present study.
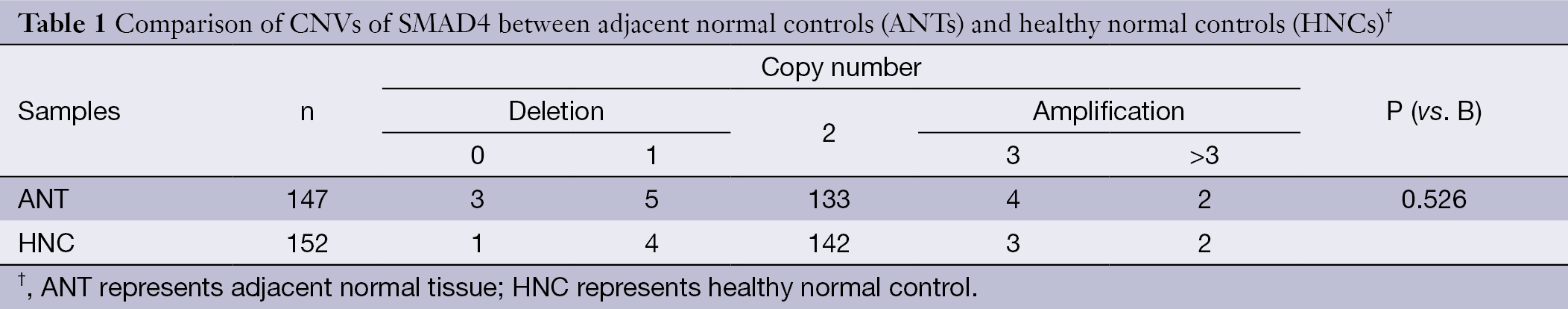
Full table
Table 2 shows CNVs of SMAD4 in paired samples of CRCs and ANTs. A total of 147 CRC samples were examined. A relatively high percentage of CRC samples showed deletions of SMAD4 (34.7%, 51 out of 147). The CRC tissues from patients with early-stage CRCs (Dukes A & B) contained on average less than 30% of the samples that had either zero or one copy of the SMAD4 gene, whereas more than 30% of samples that had either zero or one copy of SMAD4 was observed in advanced (Dukes C & D) CRCs. There is a statistical correlation between gene copy number loss and the CRC phenotypes (P<0.05).
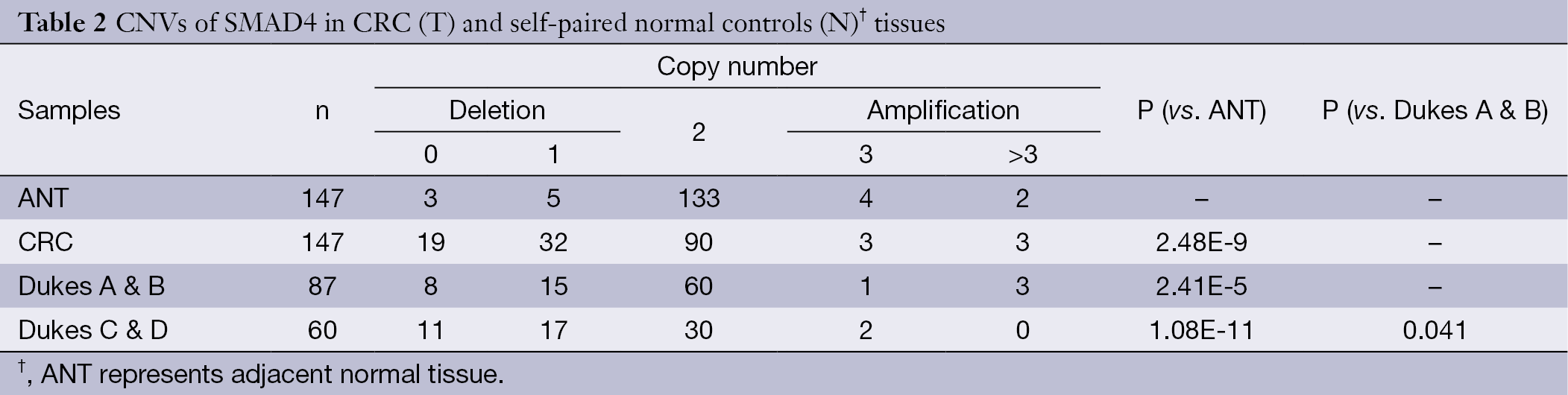
Full table
Positive correlation between copy number decreases and mRNA down-regulation of SMAD4 gene in CRCs
To find whether CNVs of SMAD4 have genotype-phenotype correlation, we compared the mRNA expression levels of SMAD4 between the CRC samples and paired ANTs by quantitative real-time RT-PCR. As shown in Figure 2A, decreased mRNA expression level of SMAD4 was observed in the CRC tissues compared with the ANTs (P<0.01). The obtained results were consistent with previous findings.

Gene CNVs can contribute to qualitative and quantitative diversities to their gene products. Next, we selected the samples with decreased or unaltered copies of SMAD4 and tested whether the SMAD4 mRNA expression was correlated with the copy numbers. The samples with increased copies of SMAD4 were not included due to the small sample size. As shown in Figure 2B, the CRC samples in the group with deleted or unaltered copies of SMAD4 both showed an impaired expression of mRNA compared with ANTs (P<0.05). There was a statistical difference between the CRC samples in the groups with deleted and unchanged copies of SMAD4 (P<0.05). Thus the DNA copy loss plays at least a partial role in the down-regulation of SMAD4 in CRCs.
Discussion
CNVs identified by CGH and array technology have been clearly shown to have the potential to directly or indirectly influence a healthy individual’s susceptibility to cancer, for example by varying the gene dosage of tumor suppressors or oncogenes. Examination of the CNVs for such genes is a starting point for investigations into the role of gene alteration in the colorectal carcinogenic process. However, there are many discrepancies among previous studies which used high-resolution approaches to screen CNVs. Thus, validation of such CNVs by a large amount of clinical samples is required.
In the present study, we collected 147 CRC samples for CNV analysis of SMAD4. Since there was no statistical difference of CNVs between the HNCs and normal tissues from CRC patients, the CNVs of SMAD4 were more likely acquired DNA aberrations in sporadic CRCs. The frequent deletion of SMAD4 (34.7%) was found in the collected CRC samples and the frequency of gene copy loss was consistent with previous studies where loss of chromosomes 18q21 were reported in 28-43% of CRCs, respectively (21-23). The results from the present study showed that the frequency of DNA copy number loss of SMAD4 in advanced CRCs was significantly more than early-stage CRC, suggesting that copy number loss played a role in CRC progression and might contribute to tumor aggressiveness. However, we found that copy numbers of SMAD4 were also increased in a small percentage of samples (6 out of 134). This discrepancy might be due to the different races and populations used in the studies. Another reason could be the different methodologies used in the different studies. We used gene-specific strategy to target short fragments (several hundred basepairs) and the sensitivity was increased.
It is expected that the CNVs do have genotype-phenotype correlation. Phenotypic effects of genetic differences, such as CNVs, are supposedly brought about by changes in expression levels (24,25). In the present study, we investigated the correlation between the SMAD4 mRNA expression and the copy numbers of its DNA. Contrary to our expectation, the correlation was not as positive as expected, although a statistical difference was obtained. Expression level of SMAD4 mRNA was increased in both the groups of deleted and unchanged DNA copies. There was a statistical difference of mRNA expression between the groups of deleted and unaltered DNA copies. Thus, CNVs did play a role of over-expression of the SMAD4 mRNA in CRCs, while there were also other mechanisms involved [e.g., site mutations (26) and CpG island hyper-methylation (27)].
Conclusions
In general, plausibly, our findings showed that the CNVs of SMAD4 have the potential to serve as diagnostic indicators, alone or in combination with other markers, for colorectal malignancies. However, the functional consequences of CNVs, the different feature of CNVs between CRCs and other gastrointestinal malignancies, and the underlying mechanisms of the heterogeneous expression levels need to be extensively investigated in the future.
Acknowledgements
This study was supported by grants from Scientific Research Foundation of Hubei Health Department (JX3B88).
Disclosure: The authors declare no conflict of interest.
References
- Sung JJ, Lau JY, Goh KL, et al. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol 2005;6:871-6. [PubMed]
- Lu JB, Sun XB, Dai DX, et al. Epidemiology of gastroenterologic cancer in Henan Province, China. World J Gastroenterol 2003;9:2400-3. [PubMed]
- Curtin K, Lin WY, George R, et al. Meta association of colorectal cancer confirms risk alleles at 8q24 and 18q21. Cancer Epidemiol Biomarkers Prev 2009;18:616-21. [PubMed]
- Salovaara R, Roth S, Loukola A, et al. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut 2002;51:56-9. [PubMed]
- Roth S, Laiho P, Salovaara R, et al. No SMAD4 hypermethylation in colorectal cancer. Br J Cancer 2000;83:1015-9. [PubMed]
- Liang JT, Chang KJ, Chen JC, et al. Hypermethylation of the p16 gene in sporadic T3N0M0 stage colorectal cancers: association with DNA replication error and shorter survival. Oncology 1999;57:149-56. [PubMed]
- Kim JH, Kang GH. Molecular and prognostic heterogeneity of microsatellite-unstable colorectal cancer. World J Gastroenterol 2014;20:4230-43. [PubMed]
- Abdel-Rahman WM, Nieminen TT, Shoman S, et al. Loss of p15INK4b expression in colorectal cancer is linked to ethnic origin. Asian Pac J Cancer Prev 2014;15:2083-7. [PubMed]
- Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996;271:350-3. [PubMed]
- Takano S, Kanai F, Jazag A, et al. Smad4 is essential for down-regulation of E-cadherin induced by TGF-beta in pancreatic cancer cell line PANC-1. J Biochem 2007;141:345-51. [PubMed]
- Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor {beta} (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol 2005;25:8108-25. [PubMed]
- Fukuchi M, Masuda N, Miyazaki T, et al. Decreased Smad4 expression in the transforming growth factor-beta signaling pathway during progression of esophageal squamous cell carcinoma. Cancer 2002;95:737-43. [PubMed]
- Horvath LG, Henshall SM, Kench JG, et al. Loss of BMP2, Smad8, and Smad4 expression in prostate cancer progression. Prostate 2004;59:234-42. [PubMed]
- Kojima K, Vickers SM, Adsay NV, et al. Inactivation of Smad4 accelerates Kras(G12D)-mediated pancreatic neoplasia. Cancer Res 2007;67:8121-30. [PubMed]
- Alazzouzi H, Alhopuro P, Salovaara R, et al. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res 2005;11:2606-11. [PubMed]
- Zhang B, Halder SK, Kashikar ND, et al. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology 2010;138:969-80.e1-3.
- Iacobuzio-Donahue CA, Song J, Parmiagiani G, et al. Missense mutations of MADH4: characterization of the mutational hot spot and functional consequences in human tumors. Clin Cancer Res 2004;10:1597-604. [PubMed]
- Barberá VM, Martín M, Mariñoso L, et al. The 18q21 region in colorectal and pancreatic cancer: independent loss of DCC and DPC4 expression. Biochim Biophys Acta 2000;1502:283-96. [PubMed]
- Akhurst RJ, Derynck R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol 2001;11:S44-51. [PubMed]
- Ghazi S, von Holst S, Picelli S, et al. Colorectal cancer susceptibility loci in a population-based study: Associations with morphological parameters. Am J Pathol 2010;177:2688-93. [PubMed]
- Lassmann S, Weis R, Makowiec F, et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med (Berl) 2007;85:293-304. [PubMed]
- Jernvall P, Mäkinen MJ, Karttunen TJ, et al. Loss of heterozygosity at 18q21 is indicative of recurrence and therefore poor prognosis in a subset of colorectal cancers. Br J Cancer 1999;79:903-8. [PubMed]
- Carethers JM, Hawn MT, Greenson JK, et al. Prognostic significance of allelic lost at chromosome 18q21 for stage II colorectal cancer. Gastroenterology 1998;114:1188-95. [PubMed]
- von Holst S, Picelli S, Edler D, et al. Association studies on 11 published colorectal cancer risk loci. Br J Cancer 2010;103:575-80. [PubMed]
- Middeldorp A, Jagmohan-Changur S, van Eijk R, et al. Enrichment of low penetrance susceptibility loci in a Dutch familial colorectal cancer cohort. Cancer Epidemiol Biomarkers Prev 2009;18:3062-7. [PubMed]
- Woodford-Richens KL, Rowan AJ, et al. SMAD4 mutations in colorectal cancer probably occur before chromosomal instability, but after divergence of the microsatellite instability pathway. Proc Natl Acad Sci U S A 2001;98:9719-23. [PubMed]
- Sameer AS, Siddiqi MA. SMAD4 promoter hypermethylation in Kashmiri colorectal cancer cases. Saudi J Gastroenterol 2011;17:297. [PubMed]
