The association of TNF-308 (G/A) gene polymorphisms and hepatocellular carcinoma risk: a meta-analysis
Qing Sun, Xuedan Guo, Qi Wang, Fan Zhao
Contributions: (I) Conception and design: Q Sun, F Zhao; (II) Administrative support: X Guo; (III) Provision of study materials or patients: X Guo; (IV) Collection and assembly of data: X Guo; (V) Data analysis and interpretation: Q Wang; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors.
Abstract
Objective: Many studies have examined the association between the TNF-308 G/A polymorphism gene polymorphisms and hepatocellular carcinoma risk in various populations, but their results have been inconsistent. To assess this relationship more precisely, a meta-analysis was performed.
Methods: The PubMed and CNKI (China National Knowledge Infrastructure) database was searched for case-control studies. Odds ratios (OR) with 95% CIs were used to determine the strength of association between the TNF-308 G/A polymorphisms and HCC risk. The pooled ORs for the risk associated with the TNF-308 G/A genotype, the A carriers (A/G + A/A) vs. the wild-type homozygotes (G/G), A/A vs. G/G were calculated, respectively. Subgroup analyses were done by ethnicity and smoking status. Heterogeneity assumptions were assessed by chi-square-based Q-test.
Results: Ultimately, 21 studies, comprising 2,923 hepatocellular carcinoma cases and 4,323 controls were included. Overall, the A carriers (G/A + A/A) vs. the wild-type homozygotes (G/G), the pooled OR was 1.05 (95% CI, 0.93-1.19; P=0.000 for heterogeneity), for A/A vs. G/G the pooled OR was 1.07 (95% CI, 0.95-1.21; P=0.007 for heterogeneity). In the stratified analysis by ethnicity, the significantly risks were found among non-Asians. However, for Asians, significantly risks were not found.
Conclusions: The TNF-308 G/A polymorphisms are not associated with hepatocellular carcinoma risk among Asians, but for non-Asians.
Keywords: TNF-308; polymorphism; hepatocellular carcinoma; susceptibility
Submitted Jul 10, 2015. Accepted for publication Jan 07, 2016.
doi: 10.21147/j.issn.1000-9604.2016.05.09
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer worldwide (748,300 new cases per year) and the third most common cause of cancer-related deaths causes (695,900 deaths per year) (1). Moreover, hepatocellular carcinoma is particularly burdensome in the Asia-Pacific region, and control policy or preventive interventions are urgently needed in this region (2, 3). Though several major risk factors of hepatocellular carcinoma have been identified, including chronic infection of hepatitis B virus and hepatitis C virus, the etiology of hepatocellular carcinoma is still unclear (4). Current studies have shown genetic factors may also contribute to the etiology of hepatocellular carcinoma, and several genetic polymorphisms have been proven to be associated with increased risk of hepatocellular carcinoma (5).
Tumor necrosis factor (TNF) is one of the cytokines principally secreted by monocyte-macrophage lineage cells, and is a ubiquitous cytokine involved in various physiologic as well as pathologic processes such as inflammation, immune-regulation, proliferation, apoptosis and oncology. In 1992, the genomic polymorphism resulting in substitution of the nucleotide adenine (A) for guanine (G) at position TNF-308 was discovered within a regulatory region of the TNF-α locus (6). The predominant homozygous allele, the heterozygous allele and the homozygous rare allele of the TNF-308 polymorphism are known as the homozygous wild-type genotype (G/G), the heterozygote (G/A) and the homozygote (A/A), respectively. Presence of the A substitution has been shown to result in increased binding of nuclear factors and enhanced transcription of the gene (7, 8).
Recently, many studies have investigated the role of the TNF-308 G/A polymorphisms in HCC. However, the results of these studies remain inconclusive. A single study might not be powered sufficiently to detect a small effect of the polymorphisms on HCC, particularly in relatively small sample sizes. Various types of study populations and study designs might also have contributed to these disparate findings. To clarify the effect of the TNF-308 G/A polymorphism on the risk for HCC, we performed a meta-analysis of all eligible case-control studies that have been published.
Materials and methods
Publication search
We searched for studies in the PubMed and CNKI (China National Knowledge Infrastructure) electronic databases to include in this meta-analysis, using the terms "liver cancer" or "hepatocellular carcinoma" "TNF-α" or "TNF" and "polymorphism". An upper date limit of March 01, 2015 was applied; no lower date limit was used. The search was performed without any restrictions on language and was focused on studies that had been conducted in humans. We also reviewed the Cochrane Library for relevant articles. Concurrently, the reference lists of reviews and retrieved articles were searched manually. Only full-text articles were included. When the same patient population appeared in several publications, only the most recent or completes study was included in this meta-analysis.
Inclusion criteria
For inclusion, the studies must have met the following criteria: they (I) evaluated TNF-308 G/A polymorphisms and HCC risk; (II) were case-control studies; (III) supplied the number of individual genotypes for the TNF-308 G/A polymorphisms in HCC cases and controls, respectively; and (IV) demonstrated that the distribution of genotypes among controls were in Hardy-Weinberg equilibrium.
Data extraction
Information was extracted carefully from all eligible publications independently by two authors, based on the inclusion criteria above. Disagreements were resolved through a discussion between the two authors.
The following data were collected from each study: first author's surname, year of publication, ethnicity, total numbers of cases and controls, and numbers of cases and controls who harbored the TNF-308 G/A genotypes, respectively. If data from any category were not reported in the primary study, the items were designated "not applicable". We did not contact the author of the primary study to request the information. Different ethnicity descents were categorized as Asian, Caucasian and mixed population. We did not require a minimum number of patients for a study to be included in our meta-analysis.
Statistical analysis
Odds ratios (OR) with 95% CIs were used to determine the strength of association between the TNF-308 G/A polymorphisms and HCC risk. The pooled ORs for the risk associated with the TNF-308 G/A genotype, the A carriers (A/G + A/A) vs. the wild-type homozygotes (G/G), A/A vs. G/G were calculated, respectively. Subgroup analyses were done by ethnicity and smoking status. Heterogeneity assumptions were assessed by chi-square-based Q-test (9). A P value greater than 0.10 for the Q-test indicated a lack of heterogeneity among the studies. Thus, the pooled OR estimate of each study was calculated using the fixed-effects model (the Mantel-Haenszel method) (10); otherwise, the random-effects model (the DerSimonian and Laird method) was used (11).
One-way sensitivity analyses were performed to determine the stability of the results-each individual study in the meta-analysis was omitted to reflect the influence of the individual dataset on the pooled OR (12). Potential publication biases were estimated by funnel plot, in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetrical plot suggests a publication bias. Funnel plot asymmetry was assessed by Egger's linear regression test, a linear regression approach that measures the funnel plot asymmetry on a natural logarithm scale of the OR. The significance of the intercept was determined by t-test, as suggested by Egger et al. (P<0.05 was considered a statistically significant publication bias) (13). All calculations were performed using STATA, version 11.0 (Stata Corporation, College Station, TX, USA).
Results
Study characteristics
A total of 21 publications involving 2,923 HCC cases and 4,323 controls met the inclusion criteria and were ultimately analyzed (14-34). Table 1 presents the main characteristics of these studies. Among the 21 publications, 20 were published in English. The sample sizes ranged from 58 to 1,245. The controls were primarily healthy populations and matched for age, ethnicity, and smoking status. There were 14 groups of Asians. All polymorphisms in the control subjects were in Hardy-Weinberg equilibrium.
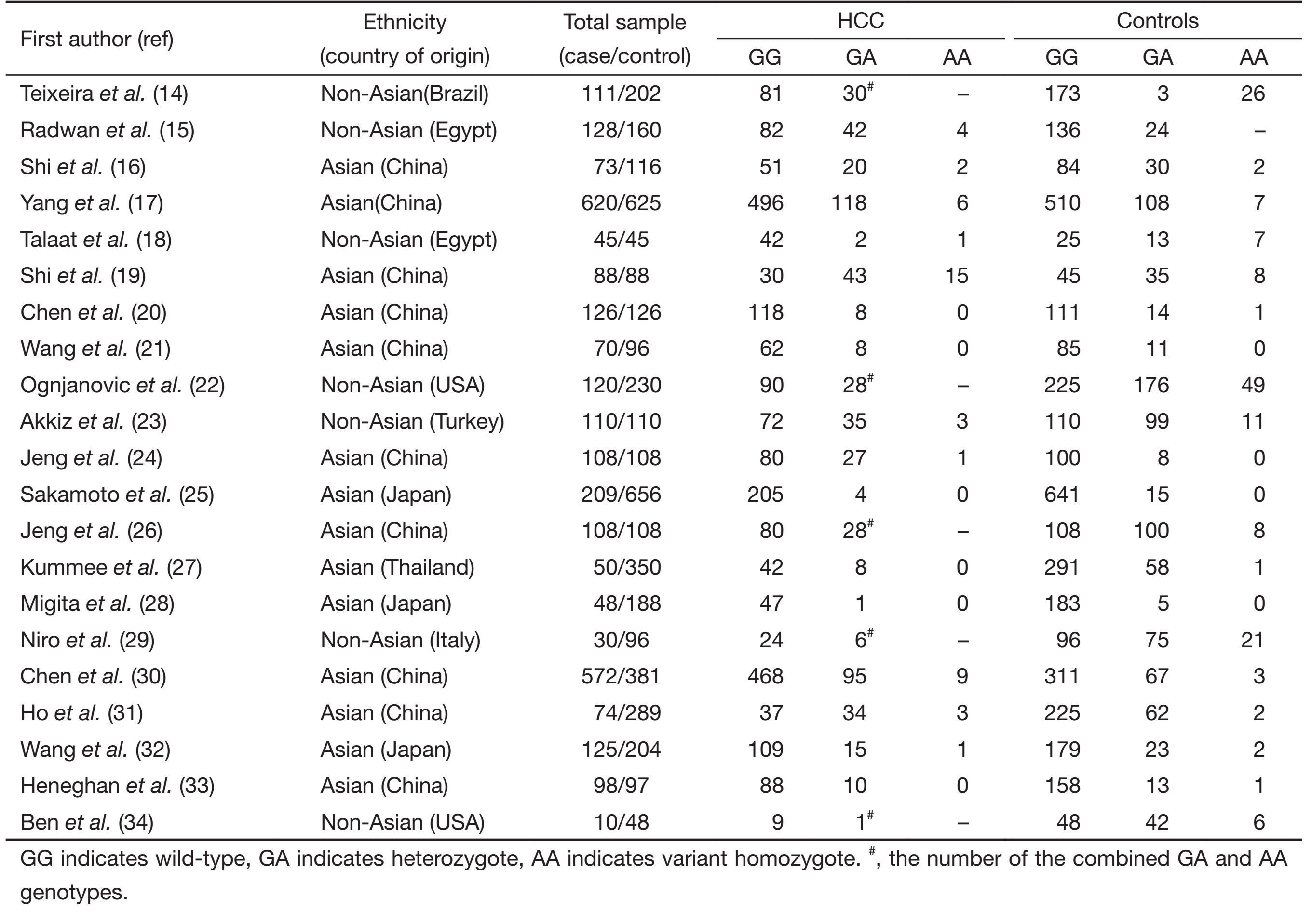
Full table
Meta-analysis results
Table 2 listed the main results of this meta-analysis. Overall, the A carriers (G/A + A/A) vs. the wild-type homozygotes (G/G), the pooled OR for all studies combined 2,923 hepatocellular carcinoma cases and 4,323 controls was 1.05 (95% CI, 0.93-1.19; P=0.000 for heterogeneity), for A/A vs. G/G the pooled OR was 1.07 (95% CI, 0.95-1.21; P=0.007 for heterogeneity) (Figure 1). In the stratified analysis by ethnicity, significantly risks were not found among Asians for (G/A + A/A) vs. (G/G) (OR =0.90, 95% CI, 0.77-1.04; P=0.000 for heterogeneity) or A/A vs. G/G (OR =0.96; 95% CI, 0.78-1.13; P=0.003 for heterogeneity). However, for non-Asians, significantly risks were found for (G/A + A/A) vs. (G/G) (OR =1.48, 95% CI, 1.19-1.85; P=0.000 for heterogeneity) and A/A vs. G/G (OR =1.44; 95% CI, 1.13-1.74; P=0.006 for heterogeneity).
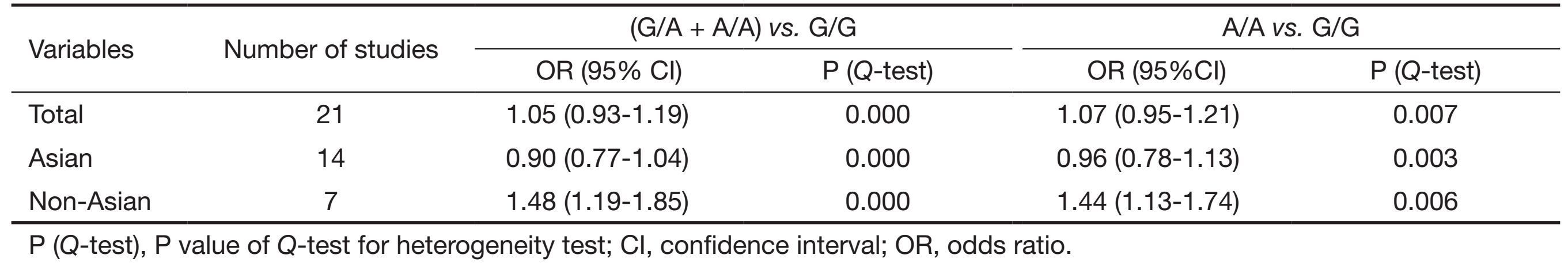
Full table
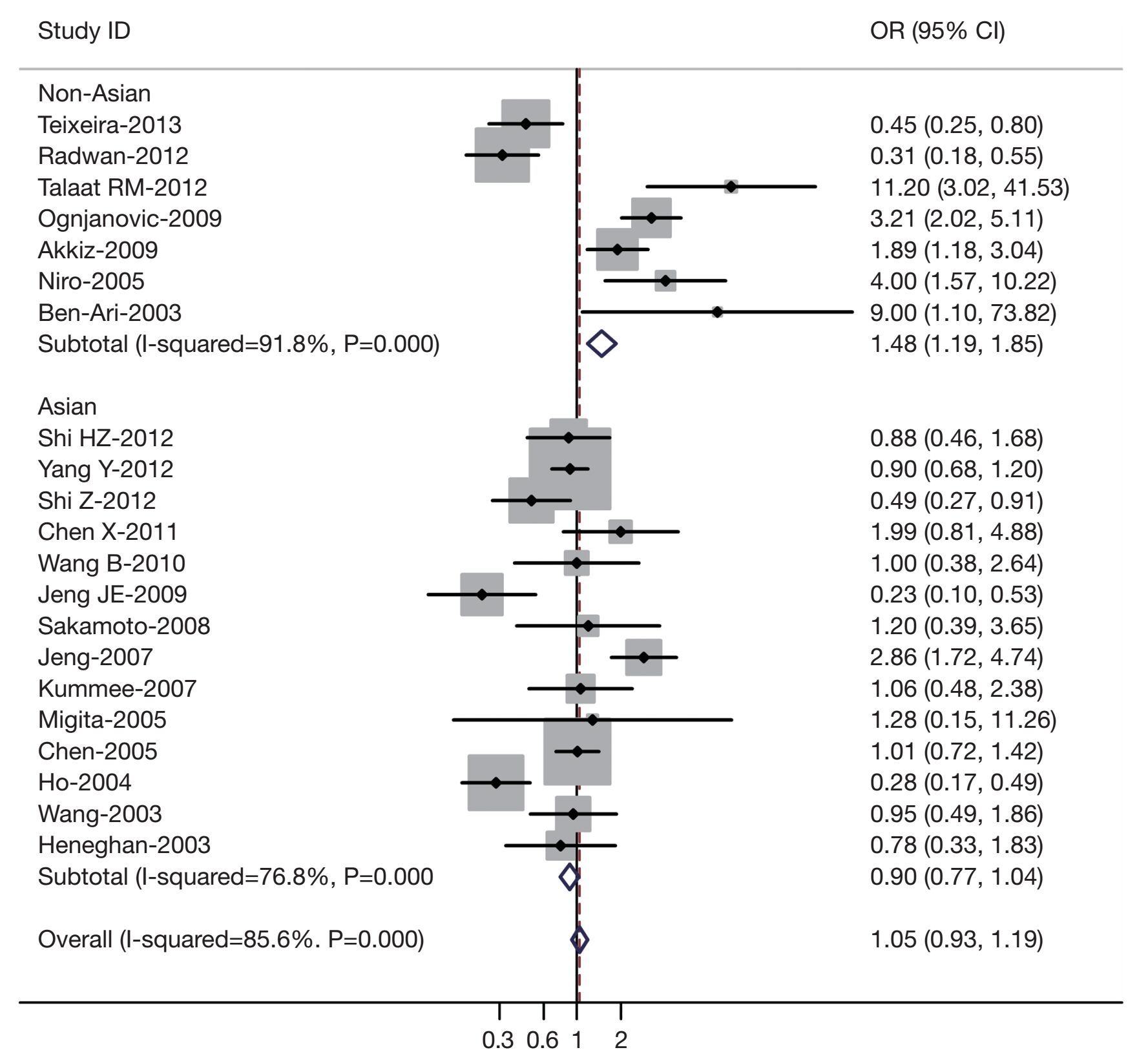
Sensitivity analyses
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled ORs, and the corresponding pooled ORs were not materially altered (data not shown).
Publication bias
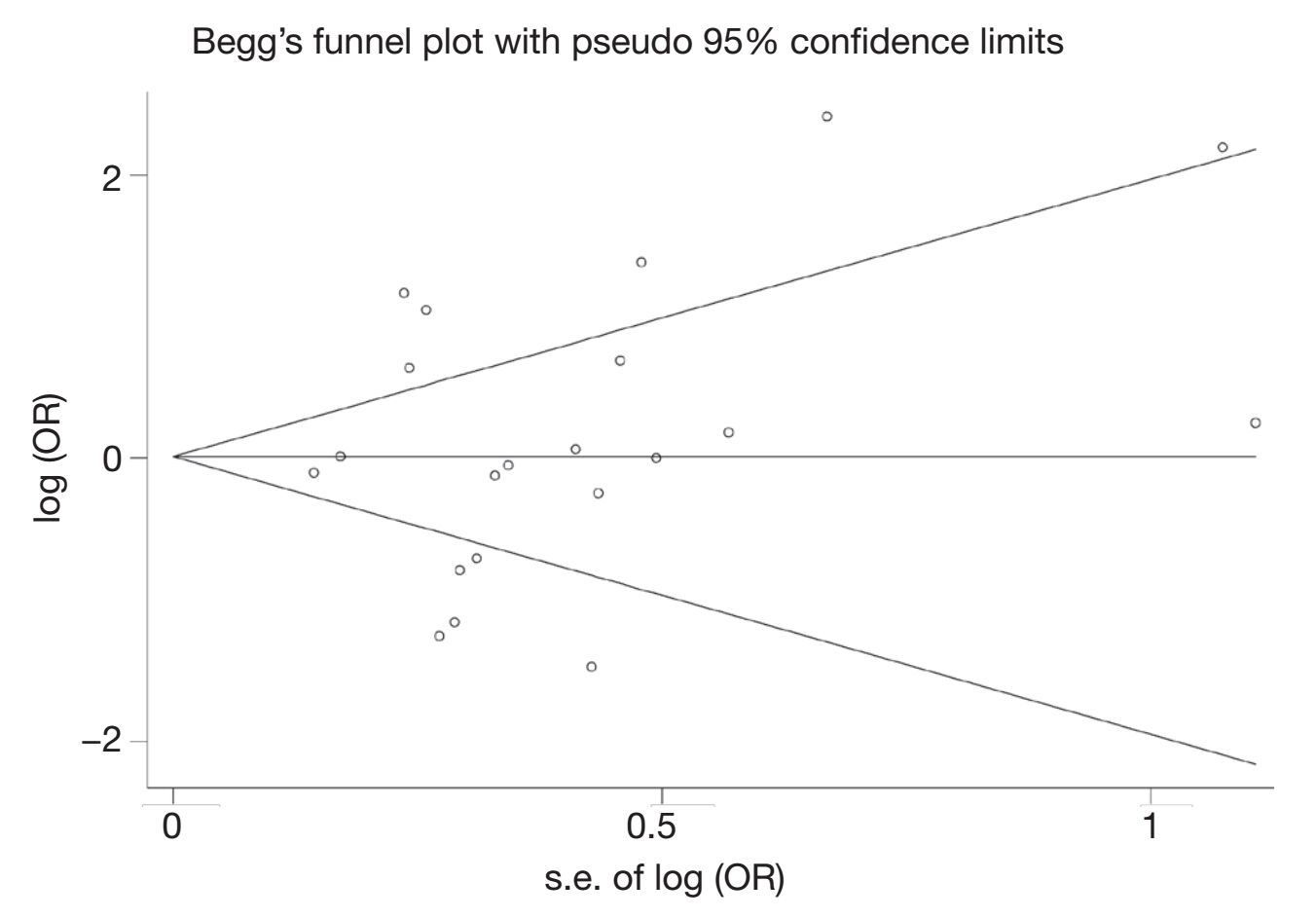
Begg's funnel plot and Egger's test were performed to access the publication bias of literatures. Evaluation of publication bias for (G/A + A/A) vs. (G/G) showed that the Egger test was significant (P=0.025). Meanwhile, for A/A vs. G/G the publication bias was found (P=0.018). The funnel plots for publication bias (Figure 2) also showed some asymmetry. These results indicated a potential for publication bias.
Discussion
Carcinogenesis of HCC is a complex, multistep and multi-factor processes, in which many factors are implicated. Although many environmental factors are found to correlate with the tumorigenesis of HCC (35, 36), the risk factors still need to be further elucidated. As we know, chronic infection with HBV or HCV is the most well established environmental risk factor for HCC worldwide. However, only a fraction of HBsAg carriers eventually develop HCC and only 2.5% of HCV infected individuals develop HCC later in life (37). Moreover, there are a portion of patients without known risk factors who eventually developed HCC (38), suggesting the inter-individual differences in susceptibility. A previous study had shown an interaction of environmental factors and genetic predisposition in the development of HCC (39). Therefore, genetic predisposition may contribute to the process of tumorigenesis.
This meta-analysis explored the association between the TNF-308 G/A gene polymorphisms and HCC risk. Our results indicated that TNF-308 G/A gene polymorphism were significantly associated with the susceptibility to HCC among non-Asians but not for Asians. However, our results were inconsistent with the previous meta-analysis (40, 41) showed that the TNF-308 G/A polymorphisms may be associated with HCC among Asians. Since then, several additional studies with large cohort populations have been reported. Simple differences in cohort populations and study design may also contribute to the disparate findings. We have improved upon that previous meta-analysis by including more recent related studies and by generally using a more comprehensive search strategy. Screening, study selection, and data extraction were performed independently and reproducibly by two reviewers. We also explored heterogeneity and potential publication bias in accordance with published guidelines.
Some limitations of this meta-analysis should be acknowledged. First, heterogeneity can interfere with the interpretation of the results of a meta-analysis. Although we minimized this likelihood by performing a careful search of published studies, using explicit criteria for a study's inclusion and performing strict data extraction and analysis, significant interstudy heterogeneity nevertheless existed in nearly every comparison. The presence of heterogeneity can result from differences in the selection of controls, age distribution, and prevalence of lifestyle factors. Although most controls were selected from healthy populations, some studies had selected controls among friends or family members of lung cancer patients or patients with other diseases. Further, only published studies were included in this meta-analysis. The presence of publication bias indicates that non-significant or negative findings might be unpublished. Finally, our results were based on unadjusted estimates; a more precise analysis should have been conducted if individual data were available, which would have allowed us to adjust using other covariates, including age, ethnicity, family history, environmental factors, and lifestyle.
Despite these limitations, this meta-analysis suggests that the TNF-308 G/A polymorphisms are not associated with HCC risk among Asians; however the gene polymorphisms are associated with non-Asians. Large-sample studies of different ethnic groups with carefully matched cases and controls should be considered in future association studies to confirm the results of our meta-analysis.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [PubMed] DOI:10.3322/caac.v61:2
- De Minicis S, Marzioni M, Benedetti A, et al. New insights in hepatocellular carcinoma: from bench to bedside. Ann Transl Med 2013;1:15. [PubMed]
- Gandhi S, Khubchandani S, Iyer R. Quality of life and hepatocellular carcinoma. J Gastrointest Oncol 2014;5:296–317. [PubMed]
- Song P, Tang W, Kokudo N. Serum biomarkers for early diagnosis of hepatocellular carcinoma. Transl Gastrointest Cancer 2014;3:103–5.
- Wang B, Huang G, Wang D, et al. Null genotypes of GSTM1 and GSTT1 contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. J Hepatol 2010;53:508–18. [PubMed] DOI:10.1016/j.jhep.2010.03.026
- Wilson AG, di Giovine FS, Blakemore AI, et al. Single base polymorphism in the human tumour necrosis factor alpha (TNF alpha) gene detectable by NcoI restriction of PCR product. Hum Mol Genet 1992;1:353.
- Kroeger KM, Carville KS, Abraham LJ. The -308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol 1997;34:391–9. [PubMed] DOI:10.1016/S0161-5890(97)00052-7
- Wilson AG, Symons JA, McDowell TL, et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A 1997;94:3195–9. [PubMed] DOI:10.1073/pnas.94.7.3195
- Cochran WG. The combination of estimates from different experiments. Biometrics 1954;10:101–29. DOI:10.2307/3001666
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [PubMed] DOI:10.1016/0197-2456(86)90046-2
- Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull 1999;8:15–7.
- Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [PubMed] DOI:10.1136/bmj.315.7109.629
- Teixeira AC, Mendes CT Jr, Marano LA, et al. Alleles and genotypes of polymorphisms of IL-18, TNF-, TNF-F-F-orphisms of IL-18, TNF-ctor alpha promoter on transcriptional activf PCR oma (HCC) in Brazil. Hum Immunol 2013;74:1024–9. [PubMed] DOI:10.1016/j.humimm.2013.04.029
- Radwan MI, Pasha HF, Mohamed RH, et al. Influence of transforming growth factor-F-F-orphisms of IL-18, TNF-ctor alpha promoter on transcriptional activf PCR oroduct. PCR product. alysis. f lifes hepatitis C patients. Cytokine 2012;60:271–6. [PubMed] DOI:10.1016/j.cyto.2012.05.010
- Shi HZ, Ren P, Lu QJ, et al. Association between EGF, TGF-β1 and TNF-α gene polymorphisms and hepatocellular carcinoma. Asian Pac J Cancer Prev 2012;13:6217–20. [PubMed] DOI:10.7314/APJCP.2012.13.12.6217
- Yang Y, Qiu X, Zeng X, et al. Correlation of polymorphism of TNF-α gene promoter with susceptibility to hepatocellular carcinoma in Guangxi. China Oncology 2012;12:3541.DOI: 10.3969/j.issn.1007-3969.2012.01.008
- Talaat RM, Esmail AA, Elwakil R, et al. Tumor necrosis factor-alpha -308G/A polymorphism and risk of hepatocellular carcinoma in hepatitis C virus-infected patients. Chin J Cancer 2012;31:29–35. [PubMed]
- Shi Z, Du C. Tumor necrosis factor alpha 308 G/A polymorphism and hepatocellular carcinoma risk in a Chinese population. Genet Test Mol Biomarkers 2011;15:569–72. [PubMed] DOI:10.1089/gtmb.2011.0008
- Chen X, Zhang L, Chang Y, et al. Association of TNF-alpha genetic polymorphisms with hepatocellular carcinoma susceptibility: a case-control study in a Han Chinese population. Int J Biol Markers 2011;26:181–187. [PubMed] DOI:10.5301/JBM.2011.8580
- Wang B, Wang J, Zheng Y, et al. A study of TNF-alpha-238 and -308 polymorphisms with different outcomes of persistent hepatitis B virus infection in China. Pathology 2010;42:674–80. [PubMed] DOI:10.3109/00313025.2010.523696
- Ognjanovic S, Yuan JM, Chaptman AK, et al. Genetic polymorphisms in the cytokine genes and risk of hepatocellular carcinoma in low-risk non-Asians of USA. Carcinogenesis 2009;30:758–62. [PubMed] DOI:10.1093/carcin/bgn286
- Akkiz H, Bayram S, Bekar A, et al. G-308A TNF-alpha polymorphism is associated with an increased risk of hepatocellular carcinoma in the Turkish population: case-control study. Cancer Epidemiol 2009;33:261–4. [PubMed] DOI:10.1016/j.canep.2009.06.001
- Jeng JE, Tsai HR, Chuang LY, et al. Independent and additive interactive effects among tumor necrosis factor-alpha polymorphisms, substance use habits, and chronic hepatitis B and hepatitis C virus infection on risk for hepatocellular carcinoma. Medicine (Baltimore) 2009;88:349–57. [PubMed] DOI:10.1097/MD.0b013e3181c10477
- Sakamoto T, Higaki Y, Hara M, et al. Interaction between interleukin-1beta -31T/C gene polymorphism and drinking and smoking habits on the risk of hepatocellular carcinoma among Japanese. Cancer Lett 2008;271:98–104. [PubMed] DOI:10.1016/j.canlet.2008.05.036
- Jeng JE, Tsai JF, Chuang LY, et al. Tumor necrosis factor-alpha 308.2 polymorphism is associated with advanced hepatic fibrosis and higher risk for hepatocellular carcinoma. Neoplasia 2007;9:987–92. [PubMed] DOI:10.1593/neo.07781
- Kummee P, Tangkijvanich P, Poovorawan Y, et al. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat 2007;14:841–8. [PubMed]
- Migita K, Miyazoe S, Maeda Y, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol 2005;42:505–10. [PubMed] DOI:10.1016/j.jhep.2004.11.026
- Niro GA, Fontana R, Gioffreda D, et al. Tumor necrosis factor gene polymorphisms and clearance or progression of hepatitis B virus infection. Liver Int 2005;25:1175–81. [PubMed] DOI:10.1111/liv.2005.25.issue-6
- Chen CC, Yang SY, Liu CJ, et al. Association of cytokine and DNA repair gene polymorphisms with hepatitis B-related hepatocellular carcinoma. Int J Epidemiol 2005;34:1310–8. [PubMed] DOI:10.1093/ije/dyi191
- Ho SY, Wang YJ, Chen HL, et al. Increased risk of developing hepatocellular carcinoma associated with carriage of the TNF2 allele of the -308 tumor necrosis factor-alpha promoter gene. Cancer Causes Control 2004;15:657–63. [PubMed] DOI:10.1023/B:CACO.0000036173.99930.75
- Wang Y, Kato N, Hoshida Y, et al. Interleukin-1beta gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology 2003;37:65–71. [PubMed] DOI:10.1053/jhep.2003.50017
- Heneghan MA, Johnson PJ, Clare M, et al. Frequency and nature of cytokine gene polymorphisms in hepatocellular carcinoma in Hong Kong Chinese. Int J Gastrointest Cancer 2003;34:19–26. [PubMed] DOI:10.1385/IJGC:34:1:19
- Ben-Ari Z, Mor E, Papo O, et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol 2003;98:144–50. [PubMed] DOI:10.1111/j.1572-0241.2003.07179.x
- Kuper H, Tzonou A, Kaklamani E, et al. Tobacco smoking, alcohol consumption and their interaction in the causation of hepatocellular carcinoma. Int J Cancer 2000;85:498–502. [PubMed] DOI:10.1002/(ISSN)1097-0215
- Chen CJ, Wang LY, Lu SN, et al. Elevated aflatoxin exposure and increased risk of hepatocellular carcinoma. Hepatology 1996;24:38–42. [PubMed] DOI:10.1002/hep.510240108
- Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 2005;436:946–52. [PubMed] DOI:10.1038/nature04079
- El-Serag HB, Mason AC. Risk factors for the rising rates of primary liver cancer in the United States. Arch Intern Med 2000;160:3227–30. [PubMed] DOI:10.1001/archinte.160.21.3227
- Yu MW, Yang SY, Chiu YH, et al. A p53 genetic polymorphism as a modulator of hepatocellular carcinoma risk in relation to chronic liver disease, familial tendency, and cigarette smoking in hepatitis B carriers. Hepatology 1999;29:697–702. [PubMed] DOI:10.1002/(ISSN)1527-3350
- Guo YM, Wei WY, Shen XZ. Tumour necrosis factor 308 polymorphisms and hepatocellular carcinoma risk: a meta-analysis. Hepatogastroenterology 2010;57:926–31. [PubMed]
- Wei Y, Liu F, Li B, et al. Polymorphisms of tumor necrosis factor-alpha and hepatocellular carcinoma risk: a HuGE systematic review and meta-analysis. Dig Dis Sci 2011;56:2227–36. [PubMed] DOI:10.1007/s10620-011-1617-y
