Remarkably different results between two studies from North America on genomic mutations and sensitivity to DNA demethylating agents for myelodysplastic syndromes
Sekeres et al. (1) conducted a multicenter randomized, controlled trial to compare whether azacitidine-based combinations with lenalidomide or vorinostat produce superior overall response rates to azacitidine in the treatment of myelodysplastic syndromes (MDS). In that trial, 224 patients with higher-risk MDS and 53 with chronic myelomonocytic leukemia (CMML) were enrolled and randomly assigned to the “azacitidine” group, “azacitidine plus lenalidomide” group or “azacitidine plus vorinostat” group. The researchers found that patients with MDS treated with azacitidine-based combinations had similar response rate to azacitidine monotherapy. Using genomic mutation analysis, they found that the overall response rate to azacitidine-based treatment was higher for patients with mutations in DNMT3A and lower for those with mutations in SRSF2. Whereas in another study, Welch et al. enrolled 26 patients with MDS and 90 with acute myeloid leukemia (AML) who were treated with decitabine, and they found that patients with TP53 mutations had a higher response rate, but not those with DNMT3A mutations (2). We propose that this big discrepancy in the conclusions between the two studies might have been caused by the presence of many co-interacting factors, e.g. study aims, DNA demethylating agents, treatment protocols, and patient sources. Still, given that both studies focused on DNA-demethylating-agent-based therapy for MDS or leukemia, it seems that a direct comparison and discussion could be fruitful.
To easily understand the response rates for patients with and without mutations of DNMT3A, SRSPF2, and TP53 obtained in the two studies, we summarized the studies’ data in Table 1. First, both studies showed a relatively higher response rate for patients with DNMT3A mutations, but only the study by Sekeres et al. showed a difference that was statistically significant. Second, both studies clearly found that patients with SRSF2 mutations showed a lower response rate. Third, the study by Welch et al. showed that patients with TP53 mutations had a much higher response rate (100%) in comparison with those with wide-type TP53 (41.0%), whereas these rates were 45.5% vs. 35.2%, respectively, in the study by Sekeres et al.
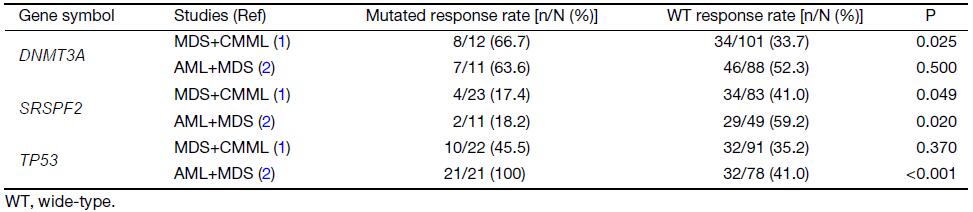
Full table
To explore the possible reason for the discrepancy between the two studies, we considered several possibilities, namely: 1) different DNA demethylating agents; 2) treatment strategies with single or combined agents; and 3) patients enriched for MDS or leukemia. However, the first two possibilities are unlikely to explain the discrepancy, because a single agent of decitabine was able to achieve a higher response rate in patients with wide-type DNMT3A or TP53 mutations than azacitidine-based combination therapy. Further, azacitidine and decitabine are antimetabolites with quite similar and unique mechanisms of action as epigenetic regulators. As for the third possibility, the patients in the studies by Sekeres et al. and Welch et al. were enriched for high-risk MDS and AML, respectively. Although Welch et al. showed that 12 AML and 9 MDS patients with TP53 mutations all showed good responses to decitabine, the number of MDS patients included was small, and unfortunately their finding could not be validated in another trial with 224 higher-risk MDS patients by Sekeres
et al.
Based on the above analysis, we might conclude that the sensitivity markers based on genomic mutations for MDS and AML might be different, although they share the same resistance marker. This finding should be further validated in the future.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Sekeres MA, Othus M, List AF, et al. Randomized phase II study of azacitidine alone or in combination with lenalidomide or with vorinostat in higher-risk myelodysplastic syndromes and chronic myelomono-cytic leukemia: North American Intergroup Study SWOG S1117. J Clin Oncol 2017;35:2745–53. DOI:10.1200/JCO.2015.66.2510
- Welch JS, Petti AA, Miller CA, et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes . N Engl J Med 2016;375:2023–36. DOI:10.1056/NEJMoa1605949
